
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,334,162 – 12,334,266 |
| Length | 104 |
| Max. P | 0.704006 |

| Location | 12,334,162 – 12,334,266 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Shannon entropy | 0.45889 |
| G+C content | 0.43455 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -8.34 |
| Energy contribution | -8.48 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
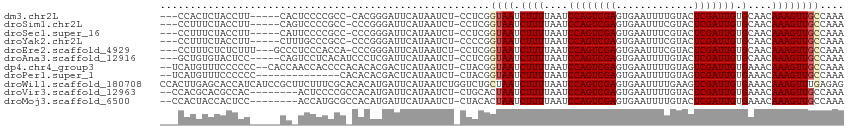
>dm3.chr2L 12334162 104 - 23011544 ---CCACUCUACCUU-----CACUCCCCGCC-CACGGGAUUCAUAAUCU-CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUACUCGAUUGUGCAACAAAGUUGCCAAA ---............-----.....((((..-..))))...........-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -25.40, z-score = -3.42, R) >droSim1.chr2L 12136272 104 - 22036055 ---CCUUUCUACCUU-----CAGUCCCCGCC-CCCGGGAUUCAUAAUCU-CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUCGUACUCGAUUGUGCAACAAAGUUGCCAAA ---............-----.((((((....-...))))))........-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -25.40, z-score = -3.48, R) >droSec1.super_16 525018 104 - 1878335 ---CCUUUCUACCUU-----CAUUCCCCGCC-CCCGGGAUUCAUAAUCU-CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUCGUACUCGAUUGUGCAACAAAGUUGCCAAA ---............-----.....((((..-..))))...........-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -25.00, z-score = -3.67, R) >droYak2.chr2L 8766777 104 - 22324452 ---CCUUUCUACCUU-----CUUUGCCCGCC-CCCGGGAUUCAUAAUCU-CCCCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUCGUACUCGAUUGUGCAACAAAGUUGCCAAA ---............-----.....((((..-..))))...........-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -24.90, z-score = -3.19, R) >droEre2.scaffold_4929 13558862 106 + 26641161 ---CCUUUCUCUCUUU---GCCCUCCCACCA-CCCGGGAUUCAUAAUCU-CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUCGUACUCGAUUGUGCAACAAAGUUGCCAAA ---.............---....((((....-...))))..........-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -24.30, z-score = -3.14, R) >droAna3.scaffold_12916 3117859 105 + 16180835 ---GCUGUGUACUCC-----CAGUCCUCACAUCCCUCGAUUCAUAAUCU-CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUACUCGAUUGUGCAACAAAGUUGCCAAA ---..((((.((...-----..))...))))..................-....(((((.((((....(((((((((((.......)))))))))).)....)))))))))... ( -21.70, z-score = -2.69, R) >dp4.chr4_group3 5921669 109 + 11692001 --UCAUGUUUCCCCCC--CACCAACCACCCCACACACGACUCAUAAUCU-CUACGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUAGUCGAUUGUGAAACAAAGUUGCCAAA --..............--...............................-....(((((.((((...(((((((((.((.......)).))))))).))...)))))))))... ( -17.60, z-score = -1.16, R) >droPer1.super_1 3024010 97 + 10282868 --UCAUGUUUCCCCCC--------------CACACACGACUCAUAAUCU-CUACGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUAGUCGAUUGUGAAACAAAGUUGCCAAA --..............--------------...................-....(((((.((((...(((((((((.((.......)).))))))).))...)))))))))... ( -17.60, z-score = -1.34, R) >droWil1.scaffold_180708 2516010 114 + 12563649 CCACUUGAGCACCAUCAUCCGCUUCUUUCGCACACAUGAUUCAUAAUCUGGUCUGCUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGAAGUCGAUUGUGAAACAAAGUUUGAGAG ...((..(((...................(((.((..(((.....)))..)).)))...........(((((((((.(.........).))))))).))......)))..)).. ( -20.60, z-score = 0.69, R) >droVir3.scaffold_12963 2775907 103 + 20206255 --CCACGCACGCCAC--------ACUCCCCGCCACAUGAUUCAUAAUCU-CUGCACUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUACUCGAUUGUGAAACAAAGUUGCCAAA --........((.((--------.......((.....(((.....))).-..)).............((((((((((((.......)))))))))).))......)).)).... ( -17.40, z-score = -1.16, R) >droMoj3.scaffold_6500 9876129 103 + 32352404 --CCACUACCACUCC--------ACCAUGCGCCACAUGAUUCAUAAUCU-CUACACUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUACUCGAUUGUGAAACAAAGUUGCCAAA --.............--------....((.((.((..(((.....))).-.................((((((((((((.......)))))))))).))......)).)))).. ( -16.90, z-score = -1.72, R) >consensus ___CCUGUCUACCUU_____CA_UCCCCGCC_CACAGGAUUCAUAAUCU_CCUCGGUAAUCUUUUAAUCCAGUCGAGUGAAUUUUGUACUCGAUUGUGCAACAAAGUUGCCAAA .......................................................((((.((((....((((((((.............))))))).)....)))))))).... ( -8.34 = -8.48 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:59 2011