| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,332,824 – 12,332,940 |
| Length | 116 |
| Max. P | 0.959047 |

| Location | 12,332,824 – 12,332,940 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 71.79 |
| Shannon entropy | 0.56113 |
| G+C content | 0.54642 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.16 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
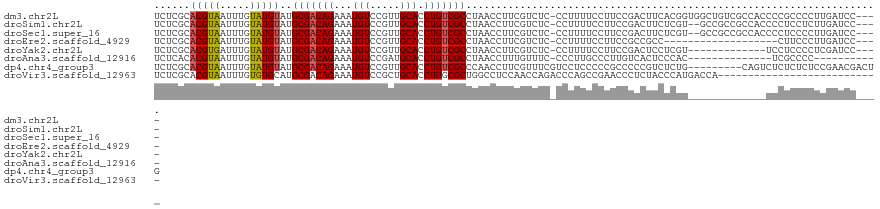
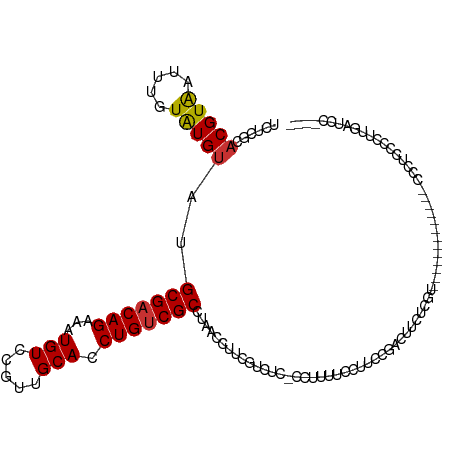
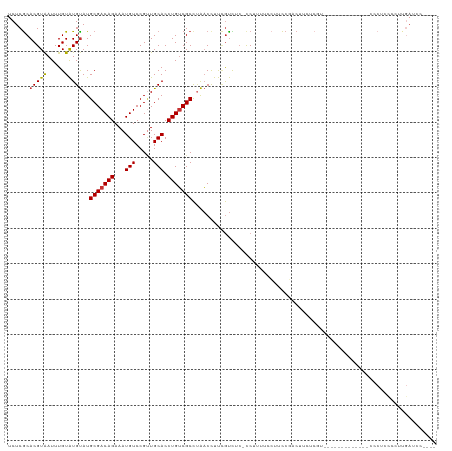
>dm3.chr2L 12332824 116 + 23011544 UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC-CCUUUUCCUUCCGACUUCACGGUGGCUGUCGCCACCCCGCCCCUUGAUCC---- ....(((((((.....)))))..(((((((...(((.....))).))))))).........(((..-............))).....((((((....))))))..))..........---- ( -25.44, z-score = -1.76, R) >droSim1.chr2L 12134796 114 + 22036055 UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC-CCUUUUCCUUCCGACUUCUCGU--GCCGCCGCCACCCCUCCUCUUGAUCC---- ....(((((..............(((((((...(((.....))).))))))).........(((..-............)))....)))--))........................---- ( -18.94, z-score = -1.75, R) >droSec1.super_16 523540 114 + 1878335 UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC-CCUUUUCCUUCCGACUUCUCGU--GCCGCCGCCACCCCUCCCCUUGAUCC---- ....(((((..............(((((((...(((.....))).))))))).........(((..-............)))....)))--))........................---- ( -18.94, z-score = -1.86, R) >droEre2.scaffold_4929 13557424 97 - 26641161 UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC-CCUUUUCCUUCCGCCGCC-------------------CUUCCCUUGAUCC---- ....(((((((.....)))))..(((((((...(((.....))).)))))))..............-............))....-------------------.............---- ( -14.40, z-score = -1.61, R) >droYak2.chr2L 8764954 103 + 22324452 UCUCGCACGUGAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC-CCUUUUCCUUCCGACUCCUCGU-------------UCCUCCCCUCGAUCC---- ..(((...(((((.((((...(((.((((....)))))))))))...))))).........(((..-............))).......-------------.........)))...---- ( -14.84, z-score = -0.79, R) >droAna3.scaffold_12916 3116542 95 - 16180835 UCUCACACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGAUGCACCUGUCGCCUAACCUUUGUUUC-CCCUUGCCCUUGUCACUCCCAC--------------UCGCCCC----------- ......(((((.....)))))..(((((((...(((.....))).)))))))..............-......................--------------.......----------- ( -12.50, z-score = -0.62, R) >dp4.chr4_group3 5920516 112 - 11692001 UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCCAACCUUCGUUUCGUCCUCCCCCGCCCCCGUCUCUG---------CAGUCUCUCUCUCCGAACGACUG ....(((((((.....)))))..(((((((...(((.....))).)))))))........................))...........---------(((((..((.....))..))))) ( -19.70, z-score = -1.21, R) >droVir3.scaffold_12963 17428192 94 + 20206255 UCUCGCACGUAAUUUGUGUGCAUGCGACAGAAAUGUCCGCUGCACCUGGCGCUGGCCUCCAACCAGACCCAGCCGAACCCUCUACCCAUGACCA--------------------------- ..(((............(((((.((((((....))).))))))))((((.(((((.......)))).))))).)))..................--------------------------- ( -26.40, z-score = -1.93, R) >consensus UCUCGCACGUAAUUUGUAUGUAUGCGACAGAAAUGUCCGUUGCACCUGUCGCCUAACCUUCGUCUC_CCUUUUCCUUCCGACUUCUCGU_____________CCCUCCCCUUGAUCC____ ......(((((.....)))))..(((((((...(((.....))).)))))))..................................................................... (-11.13 = -11.16 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:58 2011