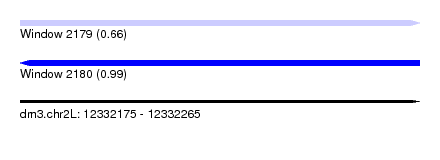
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,332,175 – 12,332,265 |
| Length | 90 |
| Max. P | 0.992693 |

| Location | 12,332,175 – 12,332,265 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Shannon entropy | 0.51347 |
| G+C content | 0.55135 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -18.19 |
| Energy contribution | -21.43 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
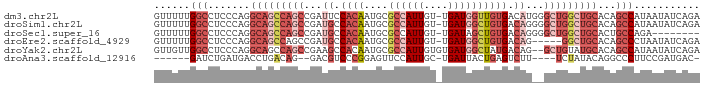
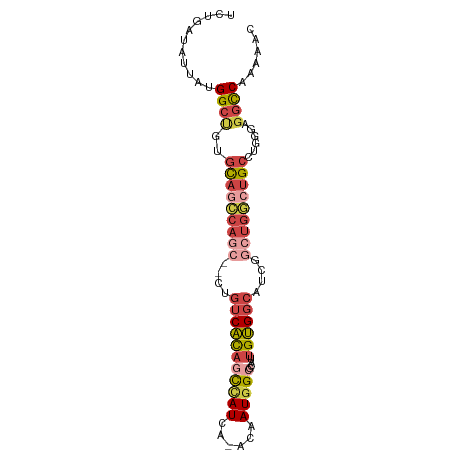
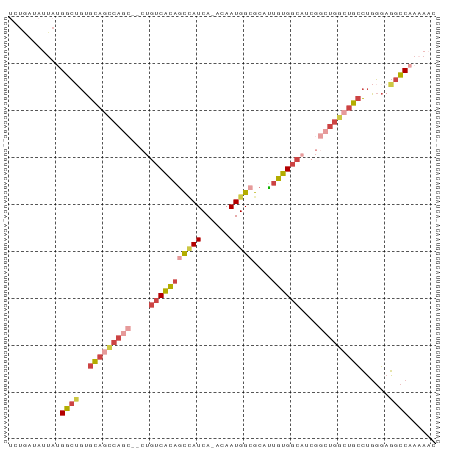
>dm3.chr2L 12332175 90 + 23011544 GUUUUUGGCCUCCCAGGCAGCCAGCCGAUUCCACAAUGCGCCAUUGU-UGAUGGUUGUGACAUGGGCUGGCUGCACAGCCAUAAUAUCAGA .....((((.......((((((((((.((..((((....((((((..-.))))))))))..)).))))))))))...)))).......... ( -37.10, z-score = -2.67, R) >droSim1.chr2L 12134118 90 + 22036055 GUUUUUGGCCUCCCAGGCAGCCAGCCGAUGCCACAAUGCGCCAUUGU-UGAUGGCUGUGACAGGGGCUGGCUGCACAGCCAUAAUAUCAGA .....((((.......((((((((((..((.((((....((((((..-.)))))))))).))..))))))))))...)))).......... ( -39.40, z-score = -2.18, R) >droSec1.super_16 522882 82 + 1878335 GUUUUUGGCCUCCCAGGCAGCCAGCCGAUGCCACAAUGCGCCAUUGU-UGAUAGCUGUGACAGGGGCUGGCUGCACUGCCAGA-------- ...((((((.....((((((((((((..((.((((..((..((....-))...)))))).))..)))))))))).))))))))-------- ( -36.80, z-score = -1.95, R) >droEre2.scaffold_4929 13556779 85 - 26641161 GUUUUUGGCCUCCCAGGCAGCCAGCCGAUGCCACAAUGCGCCAUUGU-UGAUGGCUGUGACAG-----GGCUGCACAGCCCUAAUAUCAGA (((....(((.....)))...((((((...(.((((((...))))))-.).)))))).)))((-----((((....))))))......... ( -27.70, z-score = -0.03, R) >droYak2.chr2L 8764262 89 + 22324452 GUUGUUGGCCUCCCAGGCAGCCAGCCGAAGCCACAAUGCGCCAUUGUGUGAUGGCUAUGACAG--GCUGUAUGCACAGCCAUAAUAUCAGA (((((.(((.((...(((.....))))).))))))))..((((((....)))))).......(--(((((....))))))........... ( -30.00, z-score = -0.65, R) >droAna3.scaffold_12916 3116094 77 - 16180835 ------GAUCUGAUGACCUGACAG--GACGUCCCGGAGUUCCAUUGC-UGAUUACUGAGUCUU----UCUAUACAGGCCCUUCCGAUGAC- ------.(((.((.(.((((..((--((.(.(.(((((((.......-.)))).))).).).)----)))...)))).)..)).)))...- ( -12.10, z-score = 1.67, R) >consensus GUUUUUGGCCUCCCAGGCAGCCAGCCGAUGCCACAAUGCGCCAUUGU_UGAUGGCUGUGACAG__GCUGGCUGCACAGCCAUAAUAUCAGA ......(((.......(((((((((...((.((((....((((((....)))))))))).))...)))))))))...)))........... (-18.19 = -21.43 + 3.24)

| Location | 12,332,175 – 12,332,265 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Shannon entropy | 0.51347 |
| G+C content | 0.55135 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -22.68 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.77 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
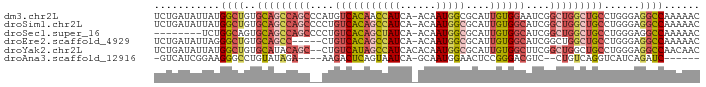
>dm3.chr2L 12332175 90 - 23011544 UCUGAUAUUAUGGCUGUGCAGCCAGCCCAUGUCACAACCAUCA-ACAAUGGCGCAUUGUGGAAUCGGCUGGCUGCCUGGGAGGCCAAAAAC ..........(((((..((((((((((.((..(((((((((..-...))))....)))))..)).))))))))))......)))))..... ( -37.00, z-score = -2.76, R) >droSim1.chr2L 12134118 90 - 22036055 UCUGAUAUUAUGGCUGUGCAGCCAGCCCCUGUCACAGCCAUCA-ACAAUGGCGCAUUGUGGCAUCGGCUGGCUGCCUGGGAGGCCAAAAAC ..........(((((..((((((((((..((((((((((((..-...)))))....)))))))..))))))))))......)))))..... ( -43.80, z-score = -3.67, R) >droSec1.super_16 522882 82 - 1878335 --------UCUGGCAGUGCAGCCAGCCCCUGUCACAGCUAUCA-ACAAUGGCGCAUUGUGGCAUCGGCUGGCUGCCUGGGAGGCCAAAAAC --------..((((((.((((((((((..((((((((((((..-...)))))....)))))))..)))))))))))).....))))..... ( -40.20, z-score = -3.06, R) >droEre2.scaffold_4929 13556779 85 + 26641161 UCUGAUAUUAGGGCUGUGCAGCC-----CUGUCACAGCCAUCA-ACAAUGGCGCAUUGUGGCAUCGGCUGGCUGCCUGGGAGGCCAAAAAC .(((((...((((((....))))-----))(((((((((((..-...)))))....))))))))))).(((((.(....).)))))..... ( -33.60, z-score = -1.34, R) >droYak2.chr2L 8764262 89 - 22324452 UCUGAUAUUAUGGCUGUGCAUACAGC--CUGUCAUAGCCAUCACACAAUGGCGCAUUGUGGCUUCGGCUGGCUGCCUGGGAGGCCAACAAC ..(((((....((((((....)))))--))))))..(((((......)))))...(((((((((((((.....)))...)))))).)))). ( -32.80, z-score = -1.25, R) >droAna3.scaffold_12916 3116094 77 + 16180835 -GUCAUCGGAAGGGCCUGUAUAGA----AAGACUCAGUAAUCA-GCAAUGGAACUCCGGGACGUC--CUGUCAGGUCAUCAGAUC------ -(((..(....)((((((.((((.----..(((((.((.....-))..(((....))).)).)))--))))))))))....))).------ ( -18.30, z-score = 0.10, R) >consensus UCUGAUAUUAUGGCUGUGCAGCCAGC__CUGUCACAGCCAUCA_ACAAUGGCGCAUUGUGGCAUCGGCUGGCUGCCUGGGAGGCCAAAAAC ...........((((..(((((((((....(((((((((((......)))))....))))))....)))))))))......))))...... (-22.68 = -24.45 + 1.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:57 2011