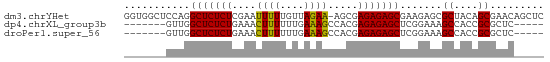
| Sequence ID | dm3.chrYHet |
|---|---|
| Location | 308,010 – 308,079 |
| Length | 69 |
| Max. P | 0.971092 |

| Location | 308,010 – 308,079 |
|---|---|
| Length | 69 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 65.66 |
| Shannon entropy | 0.44603 |
| G+C content | 0.53507 |
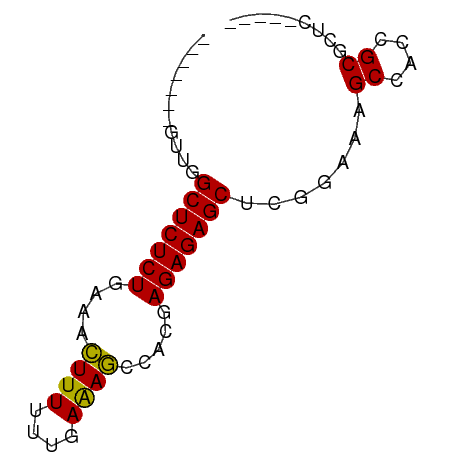
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -13.51 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
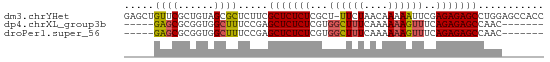
>dm3.chrYHet 308010 69 + 347038 GAGCUGUUCGCUGUAGCGCUCUUCGCUCUCUCGCU-UUCUAACAAAAAUUCGAGAGAGCCUGGAGCCACC ..((((.......))))(((((..(((((((((..-((........))..)))))))))..))))).... ( -26.60, z-score = -2.89, R) >dp4.chrXL_group3b 319160 58 - 386183 -----GAGCGCGGUGGCUUUCCGAGCUCUCUCGUGGCUUUCAAAAAAGUUUCAGAGAGCCAAC------- -----((((.(((.......))).)))).....((((((((............))))))))..------- ( -21.00, z-score = -1.76, R) >droPer1.super_56 343783 58 - 431378 -----GAGCGCGGUGGCUUUCCGAGCUCUCUCGUGGCUUUCAAAAAAGUUUCAGAGAGCCAAC------- -----((((.(((.......))).)))).....((((((((............))))))))..------- ( -21.00, z-score = -1.76, R) >consensus _____GAGCGCGGUGGCUUUCCGAGCUCUCUCGUGGCUUUCAAAAAAGUUUCAGAGAGCCAAC_______ .....((((......)))).....(((((((...((((((....))))))..)))))))........... (-13.51 = -13.73 + 0.23)
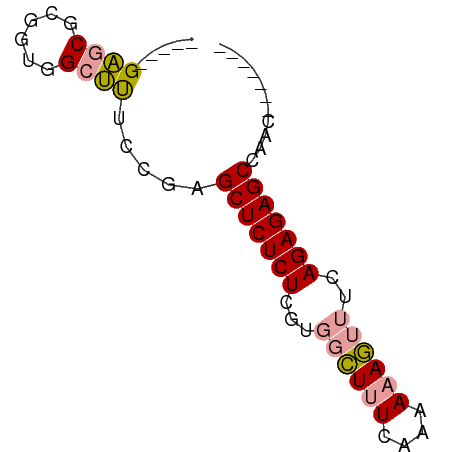
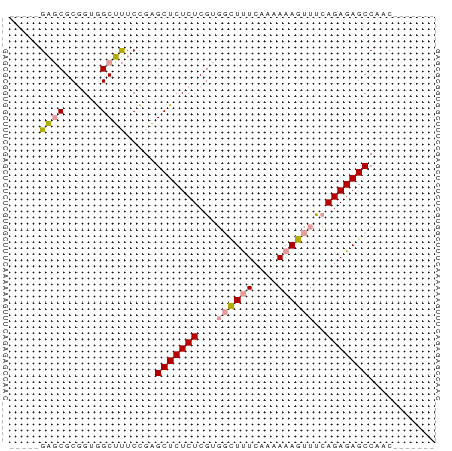
| Location | 308,010 – 308,079 |
|---|---|
| Length | 69 |
| Sequences | 3 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 65.66 |
| Shannon entropy | 0.44603 |
| G+C content | 0.53507 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -12.36 |
| Energy contribution | -11.70 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrYHet 308010 69 - 347038 GGUGGCUCCAGGCUCUCUCGAAUUUUUGUUAGAA-AGCGAGAGAGCGAAGAGCGCUACAGCGAACAGCUC (.((((((...(((((((((..((((....))))-..)))))))))...))))((....))...)).).. ( -27.20, z-score = -1.99, R) >dp4.chrXL_group3b 319160 58 + 386183 -------GUUGGCUCUCUGAAACUUUUUUGAAAGCCACGAGAGAGCUCGGAAAGCCACCGCGCUC----- -------(.(((((.(((((..((((((((.......)))))))).))))).))))).)......----- ( -18.80, z-score = -1.33, R) >droPer1.super_56 343783 58 + 431378 -------GUUGGCUCUCUGAAACUUUUUUGAAAGCCACGAGAGAGCUCGGAAAGCCACCGCGCUC----- -------(.(((((.(((((..((((((((.......)))))))).))))).))))).)......----- ( -18.80, z-score = -1.33, R) >consensus _______GUUGGCUCUCUGAAACUUUUUUGAAAGCCACGAGAGAGCUCGGAAAGCCACCGCGCUC_____ ...........(((((((....((((....)))).....))))))).......((....))......... (-12.36 = -11.70 + -0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:08:11 2011