
| Sequence ID | dm3.chrYHet |
|---|---|
| Location | 152,971 – 153,092 |
| Length | 121 |
| Max. P | 0.997018 |

| Location | 152,971 – 153,066 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.46072 |
| G+C content | 0.36898 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
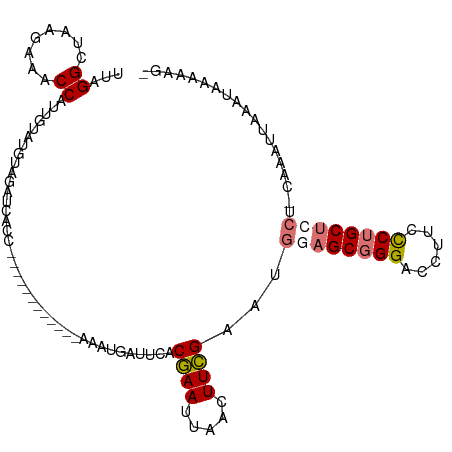
>dm3.chrYHet 152971 95 + 347038 UUAGGCUAAGAAACCAUUGUAUGUAGAUCAUA------------AGAGGAUUCACGAACUGACUUCGAAUGCAGCGGGACCUUCUCUGCUACU-UAAAUUGGAUAAAA--- ...(((..((((.((.((((((((..(((...------------....)))..))(((.....)))..)))))).))....))))..)))...-..............--- ( -16.00, z-score = 0.46, R) >droSim1.chrU 4593682 97 + 15797150 UUAGGCUAAGAAACCAUUGUAUUUAGAUCGCC------------AAAUGAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAU-AAAUAAAAAG- ...((........)).(((((.((((.(((.(------------((....))).))).))))........((((((((......)))))))))))))..-..........- ( -22.10, z-score = -2.60, R) >droSec1.super_94 39996 110 - 93855 UUAGGCUAAGAAACCAUUGUAUUUAGACCAUUGUAUUUAGAUUUAGAUUAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUCAAAUAAAAAG- ...(((((((..((....)).))))).))....(((((.(((((..........((((.....))))...((((((((......))))))))...)))))))))).....- ( -24.70, z-score = -2.55, R) >droYak2.chrU 15191482 89 - 28119190 ---GGCUAAAAAUCCUUUGUAUGGAGAACUCC------------AAGAUAUUCACAAAUUUACUUUGA-----GCGGGACCUUCCCAGCUCCA--GAAAUAAAUAAUUAAA ---..................((((....)))------------)............(((((.(((((-----(((((.....))).))))..--))).)))))....... ( -15.50, z-score = -0.67, R) >consensus UUAGGCUAAGAAACCAUUGUAUGUAGAUCACC____________AAAUGAUUCACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCU_CAAAUUAAAUAAAAAG_ ...((........)).......................................((((.....))))...((((((((......))))))))................... (-11.14 = -11.82 + 0.69)

| Location | 152,971 – 153,066 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.46072 |
| G+C content | 0.36898 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -12.43 |
| Energy contribution | -13.05 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrYHet 152971 95 - 347038 ---UUUUAUCCAAUUUA-AGUAGCAGAGAAGGUCCCGCUGCAUUCGAAGUCAGUUCGUGAAUCCUCU------------UAUGAUCUACAUACAAUGGUUUCUUAGCCUAA ---..............-.(((((((....(....).)))).......((.((.((((((......)------------))))).)))).)))...((((....))))... ( -15.20, z-score = 0.28, R) >droSim1.chrU 4593682 97 - 15797150 -CUUUUUAUUU-AUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUCAUUU------------GGCGAUCUAAAUACAAUGGUUUCUUAGCCUAA -..........-..((((((((((.(((.....))))))))........(((..(((((((....))------------)))))..))).))))).((((....))))... ( -28.90, z-score = -3.62, R) >droSec1.super_94 39996 110 + 93855 -CUUUUUAUUUGAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUAAUCUAAAUCUAAAUACAAUGGUCUAAAUACAAUGGUUUCUUAGCCUAA -.....((((((((((((((((((.(((.....))))))))...((((.....))))...................)))))..))).)))))....((((....))))... ( -24.70, z-score = -1.82, R) >droYak2.chrU 15191482 89 + 28119190 UUUAAUUAUUUAUUUC--UGGAGCUGGGAAGGUCCCGC-----UCAAAGUAAAUUUGUGAAUAUCUU------------GGAGUUCUCCAUACAAAGGAUUUUUAGCC--- ((((...((((((((.--..((((.(((.....)))))-----)).))))))))...))))...((.------------.(((((((........)))))))..))..--- ( -20.60, z-score = -0.98, R) >consensus _CUUUUUAUUUAAUUUG_AGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUAAUCU____________GAUGAUCUAAAUACAAUGGUUUCUUAGCCUAA ...................(((((.(((.....))))))))...((((.....)))).......................................((((....))))... (-12.43 = -13.05 + 0.62)



| Location | 152,996 – 153,092 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.14 |
| Shannon entropy | 0.61849 |
| G+C content | 0.36382 |
| Mean single sequence MFE | -17.45 |
| Consensus MFE | -10.76 |
| Energy contribution | -11.45 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.997018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrYHet 152996 96 + 347038 ------------GAUCAUAAGAGGAUUCACGAACUGACUUCGAAUGCAGCGGGACCUUCUCUGCUACU-UAA--AUUGGAUAAAAUAAAGAACAGCUCAAACACGAAUAAA--- ------------........(((..(((.((((.....)))).....((((((......))))))...-...--...............)))...))).............--- ( -12.00, z-score = 0.55, R) >droSim1.chrU 4593707 95 + 15797150 ------------GAUCGCCAAAUGAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCU-ACA--AAUAAAUAAAA-AGUGAAU---UCAAACUCAAAUAAUAAA ------------..((((...........((((.....))))...((((((((......)))))))).-...--...........-.))))..---.................. ( -20.60, z-score = -3.34, R) >droSec1.super_94 40021 106 - 93855 GACCAUUGUAUUUAGAUUUAGAUUAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAA--AUCAAAUAAAA-AGUGAAUA--UAAACUGAAAACCAA--- ...((((.(((((.(((((..........((((.....))))...((((((((......))))))))...))--))))))))...-))))....--...............--- ( -24.60, z-score = -3.85, R) >droYak2.chrU 15191504 94 - 28119190 ------------GAACUCCAAGAUAUUCACAAAUUUACUUUGA-----GCGGGACCUUCCCAGCUCCAGAAAUAAAUAAUUAAACAGAACGACAAACUAAAUGAGGCCUAU--- ------------(..(((...(.....)....(((((.(((((-----(((((.....))).))))..))).))))).........................)))..)...--- ( -12.60, z-score = -0.82, R) >consensus ____________GAUCUCCAAAUGAUUCACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCU_AAA__AAUAAAUAAAA_AAAGAACA__UCAAACGAAAACAAA___ .............................((((.....))))...((((((((......))))))))............................................... (-10.76 = -11.45 + 0.69)



| Location | 152,996 – 153,092 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.14 |
| Shannon entropy | 0.61849 |
| G+C content | 0.36382 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.960036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrYHet 152996 96 - 347038 ---UUUAUUCGUGUUUGAGCUGUUCUUUAUUUUAUCCAAU--UUA-AGUAGCAGAGAAGGUCCCGCUGCAUUCGAAGUCAGUUCGUGAAUCCUCUUAUGAUC------------ ---...(((((....(((((((..((((...(((......--.))-)(((((.(.(.....)).)))))....)))).))))))))))))............------------ ( -15.50, z-score = 0.36, R) >droSim1.chrU 4593707 95 - 15797150 UUUAUUAUUUGAGUUUGA---AUUCACU-UUUUAUUUAUU--UGU-AGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUCAUUUGGCGAUC------------ ........((((.(((((---((.....-...(((.....--.))-)(((((.(((.....))))))))))))))).)))).(((((((....)))))))..------------ ( -28.20, z-score = -4.06, R) >droSec1.super_94 40021 106 + 93855 ---UUGGUUUUCAGUUUA--UAUUCACU-UUUUAUUUGAU--UUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUAAUCUAAAUCUAAAUACAAUGGUC ---...............--.....(((-...((((((((--(((..(((((.(((.....))))))))...((((.....)))).........)))))).)))))....))). ( -21.90, z-score = -1.54, R) >droYak2.chrU 15191504 94 + 28119190 ---AUAGGCCUCAUUUAGUUUGUCGUUCUGUUUAAUUAUUUAUUUCUGGAGCUGGGAAGGUCCCGC-----UCAAAGUAAAUUUGUGAAUAUCUUGGAGUUC------------ ---...(..(((.....(.....)....((((((...((((((((...((((.(((.....)))))-----)).))))))))...)))))).....)))..)------------ ( -19.70, z-score = -0.68, R) >consensus ___AUUAUUCGCAUUUGA__UAUUCACU_UUUUAUUUAAU__UUU_AGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUAAUCUGAAGAUC____________ ...............................................(((((.(((.....))))))))...((((.....))))............................. (-10.90 = -11.28 + 0.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:08:08 2011