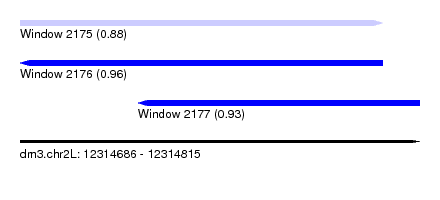
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,314,686 – 12,314,815 |
| Length | 129 |
| Max. P | 0.958632 |

| Location | 12,314,686 – 12,314,803 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Shannon entropy | 0.15879 |
| G+C content | 0.36137 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12314686 117 + 23011544 --GGGGGAAACCGAGUCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA --..((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.56, R) >droWil1.scaffold_180708 2546287 120 - 12563649 GAGAGAAACCCAAAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ....................(((((...((((((......((((....((((......)))).))))..((((((.((((....))))))))))....))))))...)))))........ ( -23.20, z-score = -1.94, R) >droPer1.super_1 2996685 117 - 10282868 --GGGGGAGACCGAGGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA --..((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.53, R) >dp4.chr4_group3 5894342 117 - 11692001 --GGGGGAGACCGAGGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA --..((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.53, R) >droAna3.scaffold_12916 3098470 117 - 16180835 --GGGGGAAACCGAGGGAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA --..((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.82, R) >droEre2.scaffold_4929 13537351 116 - 26641161 ---GGGGAAACAGCGCCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ---((.(......).))...(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -25.80, z-score = -1.84, R) >droYak2.chr2L 8745913 116 + 22324452 ---GGGGAAACCGAGUCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ---.((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.74, R) >droSec1.super_16 504342 116 + 1878335 ---GGGGAAACCGAGUCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ---.((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.74, R) >droSim1.chr2L 12115167 116 + 22036055 ---GGGGAAACCGAGUCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ---.((....))........(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -28.00, z-score = -2.74, R) >droVir3.scaffold_12963 17400820 103 + 20206255 ----------------CAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ----------------....(((((...((((((......((((....((((......)))).)))).((-((((.((((....))))))))))....))))))...)))))........ ( -23.20, z-score = -2.53, R) >droMoj3.scaffold_6500 22452773 103 + 32352404 ----------------CAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUCGAGAGCAAACCGAGCUGAAAAAACAA ----------------....(((((...((((((......((((....((((......)))).))))..(-((((.((((....))))))))).....))))))...)))))........ ( -22.00, z-score = -1.70, R) >droGri2.scaffold_15252 14837071 110 + 17193109 ---------ACCUAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA-CUGCUAAUAAGAUUAUUGCAGUCGAGAACAAACCGAGCUGAAAAAACAA ---------...........(((((...((((((......((((....((((......)))).))))..(-((((.((((....))))))))).....))))))...)))))........ ( -21.70, z-score = -1.28, R) >consensus ___GGGGAAACCGAGGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAA_CUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAA ....................(((((...((((((..(((.((((....((((......)))).))))....((((.((((....)))))))).)))..))))))...)))))........ (-20.39 = -20.48 + 0.09)

| Location | 12,314,686 – 12,314,803 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Shannon entropy | 0.15879 |
| G+C content | 0.36137 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12314686 117 - 23011544 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCCCCC-- ..(((.....(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........))).....)))............-- ( -21.00, z-score = -2.12, R) >droWil1.scaffold_180708 2546287 120 + 12563649 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUUUGGGUUUCUCUC ..........(((((((((((..(..((((((((((....)))).)))))).....)..)))))).....((((((....)))))).........((......))....)))))...... ( -21.10, z-score = -1.87, R) >droPer1.super_1 2996685 117 + 10282868 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUCUCCCCC-- ..........(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........)))....................-- ( -20.00, z-score = -2.06, R) >dp4.chr4_group3 5894342 117 + 11692001 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUCUCCCCC-- ..........(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........)))....................-- ( -20.00, z-score = -2.06, R) >droAna3.scaffold_12916 3098470 117 + 16180835 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUCCCUCGGUUUCCCCC-- ..........(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........)))....................-- ( -20.00, z-score = -2.59, R) >droEre2.scaffold_4929 13537351 116 + 26641161 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGGCGCUGUUUCCCC--- .........((((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........(((.....))))))).......--- ( -24.30, z-score = -2.84, R) >droYak2.chr2L 8745913 116 - 22324452 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCCCC--- ..(((.....(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........))).....)))...........--- ( -21.00, z-score = -2.08, R) >droSec1.super_16 504342 116 - 1878335 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCCCC--- ..(((.....(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........))).....)))...........--- ( -21.00, z-score = -2.08, R) >droSim1.chr2L 12115167 116 - 22036055 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCCCC--- ..(((.....(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........))).....)))...........--- ( -21.00, z-score = -2.08, R) >droVir3.scaffold_12963 17400820 103 - 20206255 UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUG---------------- ..........(((..((((((..(.(((((((((((....)))).))))-.)))..)..)))))).....((((((....)))))).........)))......---------------- ( -20.00, z-score = -2.89, R) >droMoj3.scaffold_6500 22452773 103 - 32352404 UUGUUUUUUCAGCUCGGUUUGCUCUCGACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUG---------------- ..........(((..((((((..(..((((((((((....)))).))))-))....)..)))))).....((((((....)))))).........)))......---------------- ( -19.50, z-score = -2.22, R) >droGri2.scaffold_15252 14837071 110 - 17193109 UUGUUUUUUCAGCUCGGUUUGUUCUCGACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGGU--------- ((((...........((((((..(..((((((((((....)))).))))-))....)..)))))).....((((((....)))))))))).((..((......))..))..--------- ( -22.00, z-score = -1.88, R) >consensus UUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG_UUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUUUCCCC___ ..........(((..((((((..(.(((((((((((....)))).))))..)))..)..)))))).....((((((....)))))).........)))...................... (-18.32 = -18.18 + -0.14)

| Location | 12,314,724 – 12,314,815 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Shannon entropy | 0.32939 |
| G+C content | 0.38963 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12314724 91 - 23011544 ---------------------GGGGCUGGGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------.(((((((((....))....)))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))............... ( -23.90, z-score = -2.90, R) >droWil1.scaffold_180708 2546327 102 + 12563649 -----------GUGGUGUGGGGGGAGAAAGGGCUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUUGUUGUUCGAACCUAAUCAAUUACCAAC -----------.(((((.(((..(((((((......)))))))..)))((((((..(..((((((((((....)))).)))))).....)..))))))........))))).. ( -26.10, z-score = -1.36, R) >droPer1.super_1 2996723 91 + 10282868 ---------------------UCGGCCUCGGUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------........(((.(((......)))...((((((..(.(((((((((((....)))).))))-.)))..)..)))))).........)))... ( -16.90, z-score = -1.64, R) >dp4.chr4_group3 5894380 91 + 11692001 ---------------------UCGGCCUCGGUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------........(((.(((......)))...((((((..(.(((((((((((....)))).))))-.)))..)..)))))).........)))... ( -16.90, z-score = -1.64, R) >droAna3.scaffold_12916 3098508 100 + 16180835 ------------GAUGCUUGUGGGCCUGGGUUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ------------(((......((((.((((..........))))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))..))).......... ( -22.00, z-score = -1.14, R) >droEre2.scaffold_4929 13537388 91 + 26641161 ---------------------GGGGCUGGGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------.(((((((((....))....)))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))............... ( -23.90, z-score = -2.90, R) >droYak2.chr2L 8745950 91 - 22324452 ---------------------GGGGCUGGGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------.(((((((((....))....)))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))............... ( -23.90, z-score = -2.90, R) >droSec1.super_16 504379 91 - 1878335 ---------------------GGGGCUGAGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------.((((((((..........))))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))............... ( -24.80, z-score = -3.38, R) >droSim1.chr2L 12115204 91 - 22036055 ---------------------GGGGCUGGGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ---------------------.(((((((((....))....)))))))((((((..(.(((((((((((....)))).))))-.)))..)..))))))............... ( -23.90, z-score = -2.90, R) >droVir3.scaffold_12963 17400844 111 - 20206255 UCUGUUUGAUUGGGAGUCUGGUCUGGUUUG-AGUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC .......(((..(....)..)))((((..(-(((((......))))))((((((..(.(((((((((((....)))).))))-.)))..)..)))))).........)))).. ( -30.00, z-score = -2.92, R) >droMoj3.scaffold_6500 22452797 112 - 32352404 UCUAUUUGAUUGGGAGUCUGUUCUGGGCUGAAGUUGUUUUUUCAGCUCGGUUUGCUCUCGACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC .....(((((((((.....((.(((((((((((......)))))))))))...))..((((..(((((((.((.......))-.))))))))))).)))))))))........ ( -35.00, z-score = -4.08, R) >droGri2.scaffold_15252 14837102 101 - 17193109 ----------UUUUGCUCUCGUCUAGUUUG-AGUUGUUUUUUCAGCUCGGUUUGUUCUCGACUGCAAUAAUCUUAUUAGCAG-UUUGUUGUUCGAACCUAAUCAAUUACCAAC ----------...................(-(((((......))))))((((((..(..((((((((((....)))).))))-))....)..))))))............... ( -23.00, z-score = -2.83, R) >consensus _____________________GGGGCUGGGCUUUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAG_UUUGUUGUUCGAACCUAAUCAAUUACCAAC ................................................((((((..(.(((((((((((....)))).))))..)))..)..))))))............... (-13.52 = -13.38 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:55 2011