| Sequence ID | dm3.chrYHet |
|---|---|
| Location | 79,970 – 80,055 |
| Length | 85 |
| Max. P | 0.999972 |

| Location | 79,970 – 80,055 |
|---|---|
| Length | 85 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 52.31 |
| Shannon entropy | 0.78791 |
| G+C content | 0.45929 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
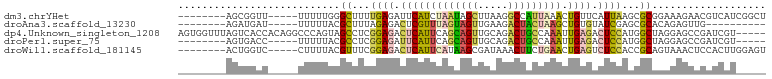
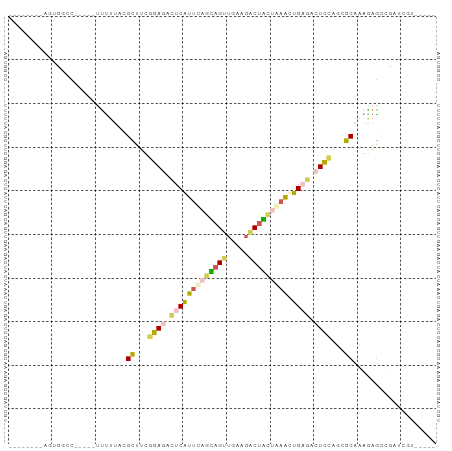
>dm3.chrYHet 79970 85 + 347038 --------AGCGGUU-----UUUUUGGGCUUUUGAGAUUCAUCUAAUAGCUUAAGGCCAUUAAACUGUUCAUUAAGCGCGGAAAGAACGUCAUCGGCU --------.(((.((-----((((((.((((.((((.......((((.((.....)).)))).....))))..)))).)))))))).)))........ ( -18.90, z-score = -0.16, R) >droAna3.scaffold_13230 681636 75 - 3602488 --------AGAUGAU-----UUUUUACGCUUUAGAGACUCGUUUAGUAGUUGAAGACUACUAAGCUGUGUAUCGAGCGCACAGAGUUG---------- --------.......-----...............((((((((((((((((...)))))))))))(((((.......)))))))))).---------- ( -21.30, z-score = -1.84, R) >dp4.Unknown_singleton_1208 2487 93 + 4803 AGUGGUUUAGUCACCACAGGCCCAGUAGCCUCGGAGACUCAUUCAGCAGUUGCAGACUGCCAAAUUGAGACUCCAUGGCUAGGAGCCGAUCGU----- .(((((......))))).(((((..(((((..((((.((((....((((((...)))))).....)))).))))..))))))).)))......----- ( -39.40, z-score = -4.33, R) >droPer1.super_75 146823 80 - 290005 --------AGUGACC-----UUUUUACGCCUCGGAGAUUCAUUCAGCAGUUGCAGACUGCCAAAUUGAGACUCCAUGGCUAGGAGCCGAUCGU----- --------.......-----((((((.(((..((((.((((....((((((...)))))).....)))).))))..)))))))))........----- ( -22.40, z-score = -1.03, R) >droWil1.scaffold_181145 855079 85 + 2682060 --------ACUGGUC-----CUUUUACGUUUCGGAGACUCAUUCAUAAGCGAUAAACUUCUGAACUGAGUCUCCACCGCAGUAAACUCCACUUGGAGU --------((((((.-----.....)).....(((((((((((((.(((.......))).)))).)))))))))....))))..(((((....))))) ( -28.20, z-score = -3.86, R) >consensus ________AGUGGCC_____UUUUUACGCUUCGGAGACUCAUUCAGCAGUUGAAGACUACUAAACUGAGACUCCAGCGCAAAGAGCCGAUCGU_____ ...........................((...((((.(((((((((((((.....))))))))).)))).))))...))................... (-12.86 = -13.38 + 0.52)

| Location | 79,970 – 80,055 |
|---|---|
| Length | 85 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 52.31 |
| Shannon entropy | 0.78791 |
| G+C content | 0.45929 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -19.14 |
| Energy contribution | -18.54 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.71 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.44 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
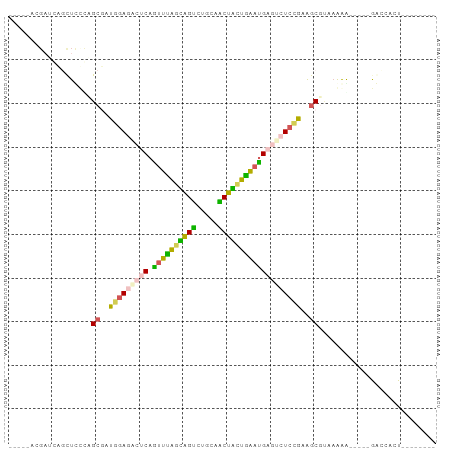
>dm3.chrYHet 79970 85 - 347038 AGCCGAUGACGUUCUUUCCGCGCUUAAUGAACAGUUUAAUGGCCUUAAGCUAUUAGAUGAAUCUCAAAAGCCCAAAAA-----AACCGCU-------- .((.((..........)).))((((...((...((((((((((.....))))))))))...))....)))).......-----.......-------- ( -15.30, z-score = -0.69, R) >droAna3.scaffold_13230 681636 75 + 3602488 ----------CAACUCUGUGCGCUCGAUACACAGCUUAGUAGUCUUCAACUACUAAACGAGUCUCUAAAGCGUAAAAA-----AUCAUCU-------- ----------........((((((.((.((...(.((((((((.....)))))))).)..)).))...))))))....-----.......-------- ( -15.80, z-score = -2.61, R) >dp4.Unknown_singleton_1208 2487 93 - 4803 -----ACGAUCGGCUCCUAGCCAUGGAGUCUCAAUUUGGCAGUCUGCAACUGCUGAAUGAGUCUCCGAGGCUACUGGGCCUGUGGUGACUAAACCACU -----......(((((.(((((.(((((.((((.((..(((((.....)))))..)))))).))))).)))))..))))).(((((......))))). ( -47.40, z-score = -6.28, R) >droPer1.super_75 146823 80 + 290005 -----ACGAUCGGCUCCUAGCCAUGGAGUCUCAAUUUGGCAGUCUGCAACUGCUGAAUGAAUCUCCGAGGCGUAAAAA-----GGUCACU-------- -----..((((........(((.(((((..(((.((..(((((.....)))))..)))))..))))).))).......-----))))...-------- ( -30.46, z-score = -3.49, R) >droWil1.scaffold_181145 855079 85 - 2682060 ACUCCAAGUGGAGUUUACUGCGGUGGAGACUCAGUUCAGAAGUUUAUCGCUUAUGAAUGAGUCUCCGAAACGUAAAAG-----GACCAGU-------- (((((....)))))....((((.((((((((((.((((.((((.....)))).))))))))))))))...))))....-----.......-------- ( -32.30, z-score = -4.19, R) >consensus _____ACGAUCAGCUCCCAGCGAUGGAGACUCAGUUUAGCAGUCUGCAACUACUGAAUGAGUCUCCGAAGCGUAAAAA_____GACCACU________ ...................((..(((((((((.((((((((((.....)))))))))))))))))))..))........................... (-19.14 = -18.54 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:08:05 2011