
| Sequence ID | dm3.chrYHet |
|---|---|
| Location | 52,634 – 52,728 |
| Length | 94 |
| Max. P | 0.537393 |

| Location | 52,634 – 52,728 |
|---|---|
| Length | 94 |
| Sequences | 15 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.64692 |
| G+C content | 0.47362 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -10.98 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrYHet 52634 94 - 347038 ---------------------ACUCUUCGUGUUUGACUUUGAGAAUACCUUCCCAGAUUCGGGAAAGAUCUCGCAGAUUAAACACAUAAUGAAAAUUGGCUGGAGUGGGUAACAU ---------------------(((((((((((((((((.(((((...(.(((((......))))).).))))).)).)))))))).((((....))))....))).))))..... ( -26.50, z-score = -2.20, R) >droSim1.chr3R 15849955 94 - 27517382 ---------------------UCACCGUGGAUAAGGUGCCAACCAUGCCCUGCCAAAUGCGCGACAAGUCGCGCAGACUGAAAACGUAGUGGAACUUGGCCGGAGUGGGCAGUAG ---------------------.((((........))))....((((..((.(((((.(((((((....))))))).((((......)))).....))))).)).))))....... ( -33.70, z-score = -1.47, R) >droSec1.super_0 16411627 94 - 21120651 ---------------------UCACCGUGGAUAAGGUGCCAACCAUGCCCUGCCAAAUGCGCGACAAGUCGCGCAGACUGAAAACGUAGUGGAACUUGGCCGGAGUGGGCAGUAG ---------------------.((((........))))....((((..((.(((((.(((((((....))))))).((((......)))).....))))).)).))))....... ( -33.70, z-score = -1.47, R) >droYak2.chr3R 19293083 94 + 28832112 ---------------------UCACCGUGGAUAAGGUGCCAACCAUGCCCUGCCAGAUGCGCGACAAAUCGCGCAGACUGAACACGUAGUGAAACUUGGCCGGAGUGGGCAGUAG ---------------------.((((........))))....((((..((.(((((.(((((((....))))))).((((......)))).....))))).)).))))....... ( -32.00, z-score = -1.17, R) >droEre2.scaffold_3327 11198 97 + 14249 ------------------UGCACUCUUCGUGUUUGACUUUGAGAAUACCUUCCCAGAUUCGAGAAAGAUCACGCAGAUUAAACACAUAAUGAAAAUUGGCUGGUGUUGGUAACAU ------------------(((..(((.(((((((((.((((.(((....))).)))).)))))......)))).)))...(((((.((((....))))....))))).))).... ( -18.40, z-score = 0.11, R) >droAna3.scaffold_13340 14148397 94 - 23697760 ---------------------UCACCGUGGAUAAGGUGCCAACCAUGCCCUGCCAGAUGCGCGACAGAUCGCGCAAACUGAAUACAUAGUGGAAUUUGGCCGGCGUGGGCAAUAG ---------------------.((((........))))....(((((((..(((((((((((((....))))))..((((......))))...))))))).)))))))....... ( -37.30, z-score = -2.78, R) >dp4.chr2 5098761 94 + 30794189 ---------------------UCACUGUGGAUAAGGUGCCAACCAUGCCCUGCCAGAUGCGCGACAGGUCGCGCAGACUGAACACGUAGUGGAACUUCGCUGGCGUGGGCAGCAG ---------------------...((((......(((....))).(((((((((((.(((((((....))))))).((((......)))).........)))))).))))))))) ( -39.70, z-score = -1.87, R) >droPer1.super_64 251803 93 + 345335 ----------------------UUCUUCUCGUUUUACUUUCAGAAUUCCUUCCCAGAUGCGUGAUAGAUCACGCAAGUUAAAAACAUAGUGAAAGUUUGAUGGUGUUGGUAACAU ----------------------......(((((..((((((((((....))).....(((((((....)))))))..............)))))))..)))))............ ( -20.10, z-score = -1.19, R) >droWil1.scaffold_181124 23006 99 + 180718 ----------------UUUACAUUCAUCUCGCUGCACCUUUAAAAUACCUUCCCAAAUACGACUGAGGUCACGCAAAUUAAAAACAUAGUGAAAAUUGGCUGGAGUGGGUAACAU ----------------...........((((((.((((.........((((.............))))((((................)))).....)).)).))))))...... ( -15.11, z-score = 0.51, R) >droVir3.scaffold_12855 4595803 94 + 10161210 ---------------------UCACUGUGGAUAAGGUGCCAACCAUGCCCUGCCAGAUGCGCGACAAGUCGCGCAGACUGAAUACGUAGUGGAAUUUGGCCGGCGUAGGCAAUAG ---------------------...............((((....(((((..(((((((((((((....))))))..((((......))))...))))))).))))).)))).... ( -32.70, z-score = -1.31, R) >droMoj3.scaffold_6501 563460 82 - 626192 ---------------------------------UUACUUUGAGAAUACCUUCCCAAAUUCGUGAAAGAUCCCUUAGAUUAAAAACGUAGUGAAAAUUAGCUGGAGUAGGUAACAU ---------------------------------............(((((.((((....(((....((((.....))))....)))((((....))))..))).).))))).... ( -13.10, z-score = -0.73, R) >droGri2.scaffold_14906 8435340 94 - 14172833 ---------------------UCACUGUGGAUAAGGUGCCAACCAUGCCUUGCCAGAUGCGCGACAAAUCGCGCAGACUGAAUACAUAGUGGAAUUUGGCCGGCGUAGGCAAUAG ---------------------((((((((..((((((.........)))))).(((.(((((((....)))))))..)))....))))))))......(((......)))..... ( -33.90, z-score = -2.36, R) >anoGam1.chr2L 429628 115 + 48795086 UUGCCGUCCGGAUGGUUUCGCACUGUUCCGAUUUGACUUUCAGCAUACCUUCCCAGAUGCGAGACAGAUCUCGCAGAUUGAACACGUAAUGGAAUUUGGCCGGUGUCGGAAGCAU ...(((.((((..(((((((.(((((((.((((((.......((((..........))))((((....)))).))))))))))).))..)))))))...))))...)))...... ( -31.50, z-score = 0.20, R) >apiMel3.GroupUn 46719617 94 - 399230636 ---------------------AUUCGGCUCUCUCAAUUCUAAGAAUACCUUCCCAAAUUCGAGAUAAAUCUCUCAGAUUAAAAACGUAAUGGAAUUUUGCCGGUGUAGGUAACAU ---------------------...((((......(((((((.((((..........))))((((......))))...............)))))))..))))((((.....)))) ( -15.20, z-score = -0.15, R) >triCas2.ChLG3 19319803 99 - 32080666 ----------------UUGGCAUUCUAGACGUUGUACUGUUAAGAUUCCUUGCCAUAUUCUUGAAAGAUCUCUCAAGUUAAACACGUAAUGGAAUUUGGCCGGUGUUGGCAACAU ----------------..............(((((((((...(((((((((((......(((((........)))))........)))).)))))))...))))....))))).. ( -20.74, z-score = -0.09, R) >consensus _____________________UCACUGUGGAUAAGAUGCUAACCAUGCCUUGCCAGAUGCGCGACAGAUCGCGCAGAUUGAAAACGUAGUGGAAUUUGGCCGGAGUGGGCAACAU .............................................(((((..((......((((....))))....((((......))))...........))...))))).... (-10.98 = -10.35 + -0.63)
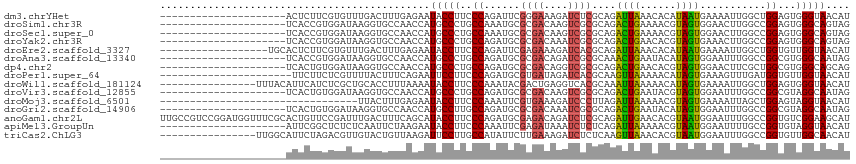
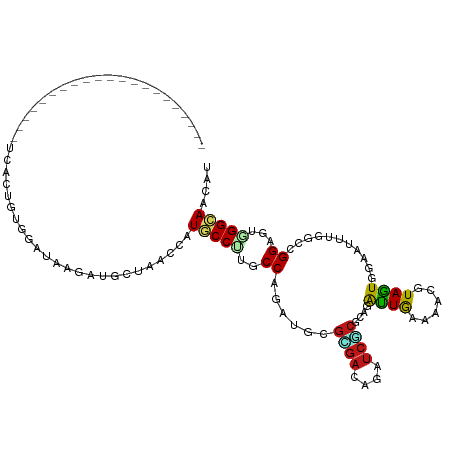
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:08:04 2011