
| Sequence ID | dm3.chrX |
|---|---|
| Location | 22,319,932 – 22,320,054 |
| Length | 122 |
| Max. P | 0.670935 |

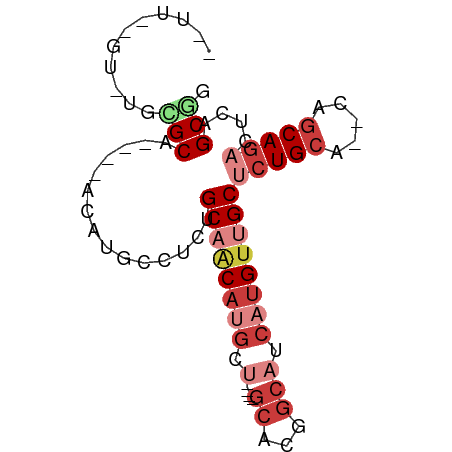
| Location | 22,319,932 – 22,320,003 |
|---|---|
| Length | 71 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.56423 |
| G+C content | 0.55611 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -11.71 |
| Energy contribution | -13.26 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
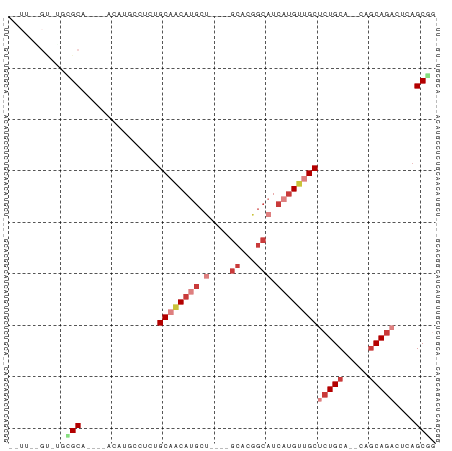
>dm3.chrX 22319932 71 + 22422827 --UUUGGUUUGAGCA----GCUUGCUUCUGCAACAUGCU----GCACGGCAUCAUGUUGCUCUGCA--CAGCAGACUCAGCGG --....(((.((((.----((((((....((((((((.(----((...))).))))))))...)))--.))).).)))))).. ( -25.50, z-score = -0.83, R) >droPer1.super_15 1597577 79 - 2181545 UUUUCUGUCGUUGCACAGAUUCAGCUUCCGCAACACGACACUAGCCCGGCAACAUGCUGCGCUGCA---CACA-ACUCAGCAA .....(((((((((..((......))...)))..)))))).((((.((((.....)))).))))..---....-......... ( -17.30, z-score = 0.06, R) >droAna3.scaffold_12929 2217199 65 - 3277472 --UUCAGCCUCAGCA----ACAUGAGUCUGCCAC------------UGGCAACAUGUUGCCCUGCAAGCUGCAGACUCAGCAA --....((....)).----...(((((((((..(------------((((((....))))).....))..))))))))).... ( -22.10, z-score = -1.26, R) >droSim1.chrU 5663249 63 - 15797150 ----------GCGCA----ACAUUCCUCUGCAACAUGCU----GCACGGCAUCAUGUAGCUCUGCA--CAGCAGACUCAGCGG ----------.(((.----..........((.(((((.(----((...))).))))).))(((((.--..)))))....))). ( -18.70, z-score = -0.74, R) >droSec1.super_39 261481 63 + 341166 ----------GCGCA----ACAUUCCUCUGCAACAUGCU----GCACGGCAUCAUGUUGCUCUGCA--CAGCAGACUCAGCGG ----------.(((.----..........((((((((.(----((...))).))))))))(((((.--..)))))....))). ( -22.80, z-score = -2.25, R) >droYak2.chrX 21677233 67 + 21770863 ------GCUUCUGCA----ACAUGCUUCUGCAGCAUGCU----GCACGGCAUCAUGUUGCUCUGCA--CAGCAGACUCAGCGG ------((((((((.----...(((....((((((((.(----((...))).))))))))...)))--..)))))...))).. ( -26.10, z-score = -1.46, R) >droEre2.scaffold_4690 18602163 71 + 18748788 --UUCUGUAGGCGCA----UCCUCGCUCUGCAACAUGCU----GCUCGGCAUCAUGUUGCUCUGCA--CAGCAGACUCAGCGG --.(((((.(((((.----.....))...((((((((.(----((...))).))))))))...)).--).)))))........ ( -23.00, z-score = -0.55, R) >consensus __UU__GU_UGCGCA____ACAUGCCUCUGCAACAUGCU____GCACGGCAUCAUGUUGCUCUGCA__CAGCAGACUCAGCGG ...........(((...............((((((((...............))))))))(((((.....)))))....))). (-11.71 = -13.26 + 1.55)

| Location | 22,319,932 – 22,320,003 |
|---|---|
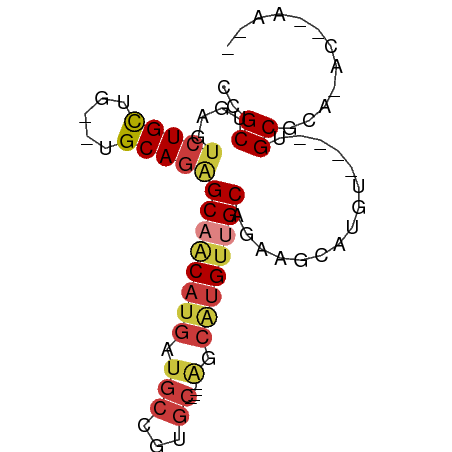
| Length | 71 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.56423 |
| G+C content | 0.55611 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.29 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 22319932 71 - 22422827 CCGCUGAGUCUGCUG--UGCAGAGCAACAUGAUGCCGUGC----AGCAUGUUGCAGAAGCAAGC----UGCUCAAACCAAA-- ....(((((..(((.--(((...((((((((.(((...))----).))))))))....))))))----.))))).......-- ( -25.40, z-score = -1.12, R) >droPer1.super_15 1597577 79 + 2181545 UUGCUGAGU-UGUG---UGCAGCGCAGCAUGUUGCCGGGCUAGUGUCGUGUUGCGGAAGCUGAAUCUGUGCAACGACAGAAAA .(((((.((-((..---..)))).)))))((((((((((.((((.(((.....)))..))))..)))).))))))........ ( -26.10, z-score = -0.06, R) >droAna3.scaffold_12929 2217199 65 + 3277472 UUGCUGAGUCUGCAGCUUGCAGGGCAACAUGUUGCCA------------GUGGCAGACUCAUGU----UGCUGAGGCUGAA-- ..(((((((((((.(((.....(((((....))))))------------)).))))))))).))----.((....))....-- ( -24.80, z-score = -0.53, R) >droSim1.chrU 5663249 63 + 15797150 CCGCUGAGUCUGCUG--UGCAGAGCUACAUGAUGCCGUGC----AGCAUGUUGCAGAGGAAUGU----UGCGC---------- ((.(((((..(((((--..(.(.((........))))..)----))))..)).))).)).....----.....---------- ( -19.40, z-score = 0.59, R) >droSec1.super_39 261481 63 - 341166 CCGCUGAGUCUGCUG--UGCAGAGCAACAUGAUGCCGUGC----AGCAUGUUGCAGAGGAAUGU----UGCGC---------- ..((....(((((..--.)))))(((((((....((.(((----((....)))))..)).))))----)))))---------- ( -23.10, z-score = -0.81, R) >droYak2.chrX 21677233 67 - 21770863 CCGCUGAGUCUGCUG--UGCAGAGCAACAUGAUGCCGUGC----AGCAUGCUGCAGAAGCAUGU----UGCAGAAGC------ ..(((...(((((..--.)))))((((((((...(..(((----((....))))))...)))))----)))...)))------ ( -25.70, z-score = -0.66, R) >droEre2.scaffold_4690 18602163 71 - 18748788 CCGCUGAGUCUGCUG--UGCAGAGCAACAUGAUGCCGAGC----AGCAUGUUGCAGAGCGAGGA----UGCGCCUACAGAA-- .((((...(((((..--.)))))((((((((.(((...))----).))))))))..))))(((.----....)))......-- ( -26.30, z-score = -1.31, R) >consensus CCGCUGAGUCUGCUG__UGCAGAGCAACAUGAUGCCGUGC____AGCAUGUUGCAGAAGCAUGU____UGCGCA_AC__AA__ ..((....(((((.....)))))((((((((...............))))))))...............))............ (-14.57 = -15.29 + 0.72)


| Location | 22,320,003 – 22,320,054 |
|---|---|
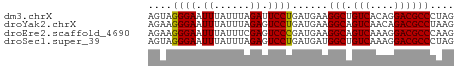
| Length | 51 |
| Sequences | 4 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Shannon entropy | 0.18608 |
| G+C content | 0.45588 |
| Mean single sequence MFE | -13.23 |
| Consensus MFE | -10.59 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 22320003 51 + 22422827 AGUAGGGAAUUUAUUUAGAUUCCUGAUGAAGGCUGUCACAGGACGCCCUAG ....((((((((....))))))))......(((.(((....)))))).... ( -13.80, z-score = -1.49, R) >droYak2.chrX 21677300 51 + 21770863 AGAAGGGAAUUUAUUUAGAGUCCUGAUGAAGGCAGUCAACAGACGCCUAAG ....((((.(((....))).)))).....((((.(((....)))))))... ( -11.80, z-score = -1.68, R) >droEre2.scaffold_4690 18602234 51 + 18748788 AGAAGGGAAUUUAUUUCGAGUCCCGAUGAAGGCAGUCAAAGGACGCCCAAG ....((((.((......)).))))......(((.(((....)))))).... ( -11.90, z-score = -1.04, R) >droSec1.super_39 261544 51 + 341166 AGUAGGGAAUUUAUUUAGAGUCCUGAUGAUGGCUGUCAAAGGACGCCCUAG ..(((((............(((((..((((....)))).))))).))))). ( -15.42, z-score = -2.16, R) >consensus AGAAGGGAAUUUAUUUAGAGUCCUGAUGAAGGCAGUCAAAGGACGCCCAAG ....((((.((......)).))))......(((.(((....)))))).... (-10.59 = -10.40 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:58 2011