
| Sequence ID | dm3.chrX |
|---|---|
| Location | 22,264,156 – 22,264,229 |
| Length | 73 |
| Max. P | 0.833503 |

| Location | 22,264,156 – 22,264,229 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.47586 |
| G+C content | 0.45322 |
| Mean single sequence MFE | -15.65 |
| Consensus MFE | -7.54 |
| Energy contribution | -7.02 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
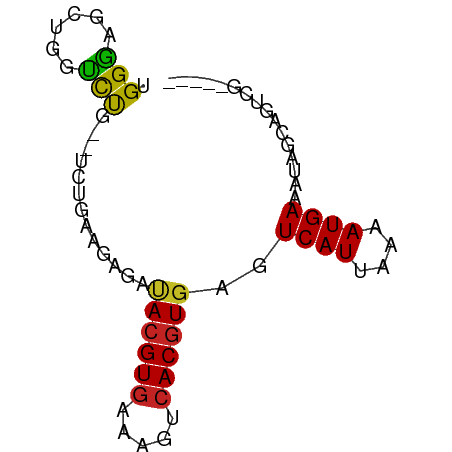
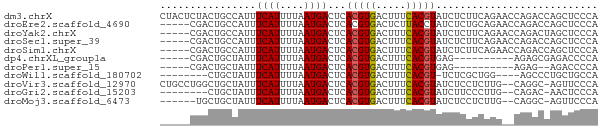
>dm3.chrX 22264156 73 + 22422827 UGGGAGCUGGUCUGGUUCUGAAGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUGGCAGUAGAGUAG (.((((((.....)))))).).....((((((.....)))))).((((((.......)))))).......... ( -17.10, z-score = -1.78, R) >droEre2.scaffold_4690 18546608 68 + 18748788 UGGGAGCUGGUCUGGUUCUGCAGAGAUAGGUAAGAGUCACGUGAGUCAUUAAAAUGAAAUGGCAGUCG----- ..((..((..((((.(((....))).))))..))..))......((((((.......)))))).....----- ( -11.30, z-score = 0.91, R) >droYak2.chrX 21627946 68 + 21770863 UGGGAGCUAGUCUGGUUCUGAAGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUGGCAGUCG----- (.((((((.....)))))).)...((((((((.....)))))..((((((.......)))))).))).----- ( -17.30, z-score = -1.77, R) >droSec1.super_39 215532 68 + 341166 UGGGAGCUGGUCUGGUUCUGAAGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUGGCAGUCG----- (.((((((.....)))))).)...((((((((.....)))))..((((((.......)))))).))).----- ( -17.50, z-score = -1.73, R) >droSim1.chrX 17008550 68 + 17042790 UGGGAGCUGGUCUGGUUCUGAAGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUGGCAGUCG----- (.((((((.....)))))).)...((((((((.....)))))..((((((.......)))))).))).----- ( -17.50, z-score = -1.73, R) >dp4.chrXL_group1a 881972 58 - 9151740 UGGGGUCUCGCUCU----------CUCACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAGUCG----- ....(.((.(((..----------((((((((.....))))))))((((....))))...))))).).----- ( -17.00, z-score = -2.83, R) >droPer1.super_15 1541733 56 - 2181545 UGGGGUCU--CUCU----------CUCACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAGUCG----- .(((....--))).----------((((((((.....))))))))((((....))))...........----- ( -13.90, z-score = -2.01, R) >droWil1.scaffold_180702 2505431 60 - 4511350 UGGCAGCAGGGCU----CCAGCGAGA-ACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAG-------- (((.(((...)))----)))((....-(((((.....)))))...((((....))))....))..-------- ( -13.40, z-score = -1.02, R) >droVir3.scaffold_12970 4532089 70 + 11907090 UGGGAACU-GCCUG--CAAGAGGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAGCCAGGCAG ......((-(((((--(.........((((((.....))))))..((((....)))).......).))))))) ( -20.30, z-score = -2.76, R) >droGri2.scaffold_15203 1246229 62 - 11997470 UGGGAGUU-GUCUG--CAAGGGAAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAG-------- ........-..(((--(.........((((((.....))))))..((((....))))....))))-------- ( -12.80, z-score = -1.85, R) >droMoj3.scaffold_6473 7515878 64 + 16943266 UGGGAACU-GCCUG--CAAGAGGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAGCA------ ........-((.((--(.........((((((.....))))))..((((....))))....))))).------ ( -14.00, z-score = -1.24, R) >consensus UGGGAGCUGGUCUG__UCUGAAGAGAUACGUGAAAGUCACGUGAGUCAUUAAAAUGAAAUAGCAGUCG_____ .(((......))).............((((((.....))))))..((((....))))................ ( -7.54 = -7.02 + -0.52)


| Location | 22,264,156 – 22,264,229 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.47586 |
| G+C content | 0.45322 |
| Mean single sequence MFE | -11.54 |
| Consensus MFE | -5.65 |
| Energy contribution | -5.65 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 22264156 73 - 22422827 CUACUCUACUGCCAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCUUCAGAACCAGACCAGCUCCCA ........(((.....((((....))))...(((((.....))))).............)))........... ( -8.30, z-score = -1.02, R) >droEre2.scaffold_4690 18546608 68 - 18748788 -----CGACUGCCAUUUCAUUUUAAUGACUCACGUGACUCUUACCUAUCUCUGCAGAACCAGACCAGCUCCCA -----...((((....((((....)))).....((((...))))........))))................. ( -5.30, z-score = 0.55, R) >droYak2.chrX 21627946 68 - 21770863 -----CGACUGCCAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCUUCAGAACCAGACUAGCUCCCA -----...(((.....((((....))))...(((((.....))))).............)))........... ( -8.30, z-score = -0.48, R) >droSec1.super_39 215532 68 - 341166 -----CGACUGCCAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCUUCAGAACCAGACCAGCUCCCA -----.(((((.....((((....))))...(((((.....)))))..................))).))... ( -8.60, z-score = -0.69, R) >droSim1.chrX 17008550 68 - 17042790 -----CGACUGCCAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCUUCAGAACCAGACCAGCUCCCA -----.(((((.....((((....))))...(((((.....)))))..................))).))... ( -8.60, z-score = -0.69, R) >dp4.chrXL_group1a 881972 58 + 9151740 -----CGACUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUGAG----------AGAGCGAGACCCCA -----...(((((...((((....))))((((((((.....))))))))----------..))).))...... ( -17.60, z-score = -4.05, R) >droPer1.super_15 1541733 56 + 2181545 -----CGACUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUGAG----------AGAG--AGACCCCA -----...((.((...((((....))))((((((((.....))))))))----------))))--........ ( -14.50, z-score = -3.07, R) >droWil1.scaffold_180702 2505431 60 + 4511350 --------CUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGU-UCUCGCUGG----AGCCCUGCUGCCA --------..((....((((....))))...(((((.....)))))-....)).((----(((...))).)). ( -10.70, z-score = -0.87, R) >droVir3.scaffold_12970 4532089 70 - 11907090 CUGCCUGGCUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCCUCUUG--CAGGC-AGUUCCCA (((((((.........((((....))))...(((((.....)))))...........--)))))-))...... ( -19.00, z-score = -3.32, R) >droGri2.scaffold_15203 1246229 62 + 11997470 --------CUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUUCCCUUG--CAGAC-AACUCCCA --------((((....((((....))))...(((((.....)))))..........)--)))..-........ ( -11.70, z-score = -3.93, R) >droMoj3.scaffold_6473 7515878 64 - 16943266 ------UGCUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCCUCUUG--CAGGC-AGUUCCCA ------.((((((...((((....))))...(((((.....)))))...........--..)))-)))..... ( -14.30, z-score = -2.64, R) >consensus _____CGACUGCUAUUUCAUUUUAAUGACUCACGUGACUUUCACGUAUCUCUUCAGA__CAGACCAGCUCCCA ................((((....))))...(((((.....)))))........................... ( -5.65 = -5.65 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:51 2011