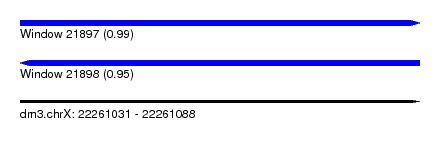
| Sequence ID | dm3.chrX |
|---|---|
| Location | 22,261,031 – 22,261,088 |
| Length | 57 |
| Max. P | 0.992522 |

| Location | 22,261,031 – 22,261,088 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Shannon entropy | 0.39719 |
| G+C content | 0.38719 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -10.40 |
| Energy contribution | -11.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
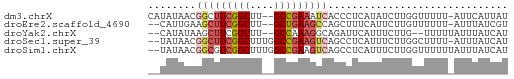
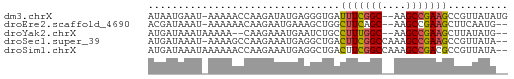
>dm3.chrX 22261031 57 + 22422827 CAUAUAACGGCUUCGGCUU--GCCGAAAUCACCCUCAUAUCUUGGUUUUU-AUUCAUUAU ........(..(((((...--.)))))..)....................-......... ( -8.60, z-score = -0.95, R) >droEre2.scaffold_4690 18543596 55 + 18748788 --CAUUGAAGCUUCGGCUU--GCUGAAGCCAGCUUUCAUUCUUGUUUUUU-AUUUAUCGU --....((((((..(((((--....)))))))))))..............-......... ( -11.10, z-score = -1.05, R) >droYak2.chrX 21624917 54 + 21770863 --CAUAUAAGCUUCGGCUU--GCCAAAGGCAGAUUCAUUUCUUG--UUUUUAUUUAUCAU --....(((((....))))--)..(((((((((......)).))--)))))......... ( -11.30, z-score = -2.35, R) >droSec1.super_39 212509 57 + 341166 --UAUAACGGCUUCGGCUUUGGCCGAAGUCAGCCUCAUUUCUUGGCUUUU-AUUUAUCAU --......(((((((((....)))))))))((((.........))))...-......... ( -21.00, z-score = -4.92, R) >droSim1.chrX 17004993 58 + 17042790 --UAUAACGGCGUCGGCUUUGGCCGAAGUCAGCCUCAUUUCUUGGUUUUUUAUUUAUCAU --......(((.(((((....))))).)))((((.........))))............. ( -14.50, z-score = -2.27, R) >consensus __UAUAACGGCUUCGGCUU__GCCGAAGUCAGCCUCAUUUCUUGGUUUUU_AUUUAUCAU ........(((((((((....))))))))).............................. (-10.40 = -11.32 + 0.92)
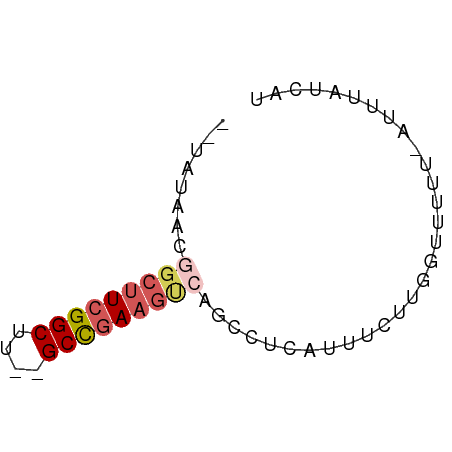
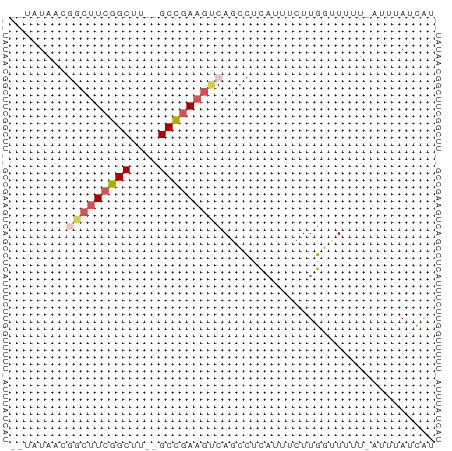
| Location | 22,261,031 – 22,261,088 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 76.86 |
| Shannon entropy | 0.39719 |
| G+C content | 0.38719 |
| Mean single sequence MFE | -12.06 |
| Consensus MFE | -7.44 |
| Energy contribution | -7.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
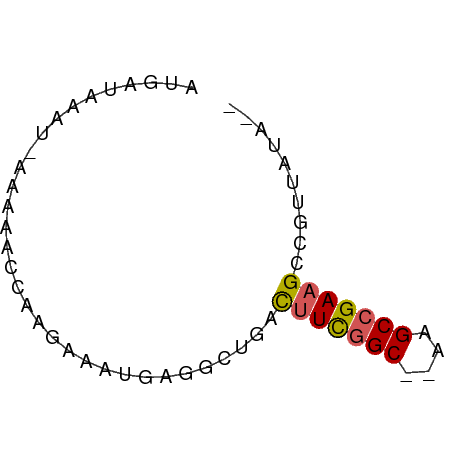
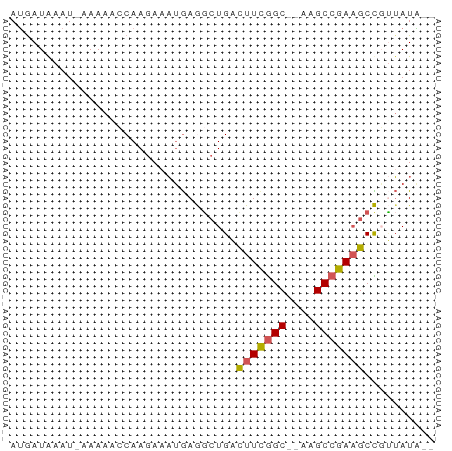
>dm3.chrX 22261031 57 - 22422827 AUAAUGAAU-AAAAACCAAGAUAUGAGGGUGAUUUCGGC--AAGCCGAAGCCGUUAUAUG .........-..........((((((.(((...(((((.--...)))))))).)))))). ( -14.80, z-score = -3.06, R) >droEre2.scaffold_4690 18543596 55 - 18748788 ACGAUAAAU-AAAAAACAAGAAUGAAAGCUGGCUUCAGC--AAGCCGAAGCUUCAAUG-- .........-............((((.((((((((....--)))))..)))))))...-- ( -10.70, z-score = -1.69, R) >droYak2.chrX 21624917 54 - 21770863 AUGAUAAAUAAAAA--CAAGAAAUGAAUCUGCCUUUGGC--AAGCCGAAGCUUAUAUG-- ..............--..............((.(((((.--...))))))).......-- ( -6.60, z-score = -0.57, R) >droSec1.super_39 212509 57 - 341166 AUGAUAAAU-AAAAGCCAAGAAAUGAGGCUGACUUCGGCCAAAGCCGAAGCCGUUAUA-- (((((....-...((((.(....)..))))(.(((((((....))))))).)))))).-- ( -17.10, z-score = -3.62, R) >droSim1.chrX 17004993 58 - 17042790 AUGAUAAAUAAAAAACCAAGAAAUGAGGCUGACUUCGGCCAAAGCCGACGCCGUUAUA-- ......................(((((((.....(((((....))))).))).)))).-- ( -11.10, z-score = -1.60, R) >consensus AUGAUAAAU_AAAAACCAAGAAAUGAGGCUGACUUCGGC__AAGCCGAAGCCGUUAUA__ ................................(((((((....))))))).......... ( -7.44 = -7.52 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:50 2011