
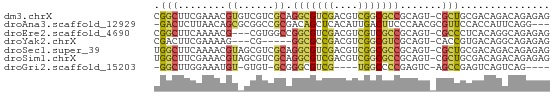
| Sequence ID | dm3.chrX |
|---|---|
| Location | 22,257,030 – 22,257,095 |
| Length | 65 |
| Max. P | 0.597745 |

| Location | 22,257,030 – 22,257,095 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 64.44 |
| Shannon entropy | 0.71703 |
| G+C content | 0.65337 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -9.83 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
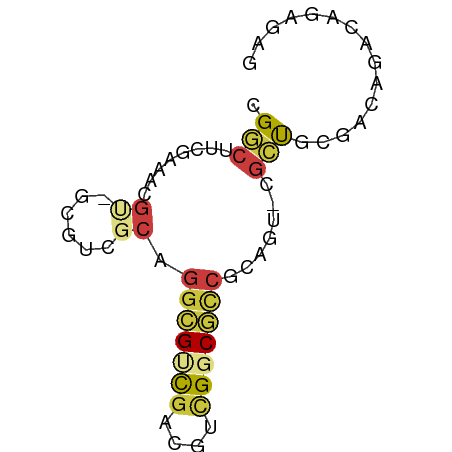
>dm3.chrX 22257030 65 + 22422827 CGGCUUCGAAACGUGUCGUCGCAGGCGUCGACGUCGGCGCCGCAGU-CGCUGCGACAGACAGAGAG .............(((((((((((((((((....)))))))((...-.)))))))).))))..... ( -28.80, z-score = -0.90, R) >droAna3.scaffold_12929 2170104 62 - 3277472 -GACUCUUAACAGCGCGGCCGCGACAGCUCACAUUGACUUCCCAACGCGUUCCACCAUUCAGG--- -..........((((((...((....)).....(((......)))))))))............--- ( -9.30, z-score = 0.33, R) >droEre2.scaffold_4690 18539965 62 + 18748788 CGGCUUCAAAACG---CGUGGCCGGCGUCGACGUCGUCGCCGCAGU-CGCCCUCACAGGCAGAGAG ..((((......(---((..((.((((.((....)).))))))...-)))......))))...... ( -20.40, z-score = 0.51, R) >droYak2.chrX 21621032 57 + 21770863 CGACUUCGAAAAG---CG-----GGCGCCGACGUCGGCGUCGCAGU-CACCGUGACAGGCAGAGAG ...((..(....(---((-----(.(((((....))))))))).((-(.....)))...)..)).. ( -19.40, z-score = -0.12, R) >droSec1.super_39 208632 65 + 341166 UGGCUUCAAAACGUAGCGUCGCAGGCGUCGACGUCGGCGCCGCAGU-CGCUGCGACAGACAGAGAG ...........(((((((.(((.(((((((....))))))))).).-)))))))............ ( -28.70, z-score = -1.81, R) >droSim1.chrX 17001230 65 + 17042790 UGGCUUCGAAACGUAGCGUCGCAGGCGUCGACGUCGGCGCCGCAGU-CGCUGCGACAGACAGAGAG ...........(((((((.(((.(((((((....))))))))).).-)))))))............ ( -28.70, z-score = -1.23, R) >droGri2.scaffold_15203 1237475 54 - 11997470 -GGCUUGGAAAUGU-GUGU-GCGGGCGUCG----UGGCCCCGAGUC-AGCCGAGUCAGUCAG---- -(((((((...((.-...(-(.((((....----..))))))...)-).)))))))......---- ( -20.20, z-score = -1.59, R) >consensus CGGCUUCGAAACGU_GCGUCGCAGGCGUCGACGUCGGCGCCGCAGU_CGCUGCGACAGACAGAGAG .(((........((......)).(((((((....))))))).......)))............... ( -9.83 = -10.16 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:48 2011