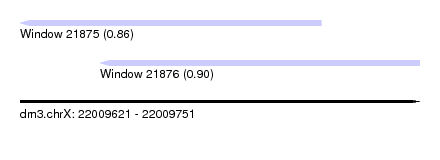
| Sequence ID | dm3.chrX |
|---|---|
| Location | 22,009,621 – 22,009,751 |
| Length | 130 |
| Max. P | 0.895307 |

| Location | 22,009,621 – 22,009,719 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.81 |
| Shannon entropy | 0.56466 |
| G+C content | 0.43726 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 22009621 98 - 22422827 -------CCGCCAUCCAGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGGCGCAUUAGGCAAAAAUUAUCAACGCACACAGAAGAGCAAAGGAAAAAA------------- -------(((((...((((..........))))..........((((.........))))....))).............((.(......).))...))......------------- ( -17.50, z-score = 0.51, R) >droSec1.super_55 162853 98 - 187931 -------CCGCCAUCCAGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAAAAAUUAUCAACGCACACAGAAGAGCAAAGGAAAAAA------------- -------(((((...((((..........))))...((((.((((....)))).))))......))).............((.(......).))...))......------------- ( -21.20, z-score = -1.23, R) >droYak2.chrX 21430435 101 - 21770863 ----UUCCAGCUCUUCGGAUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAAAAAUUAUCAACGCACACAGAAGUGCAAAUGGAAAAA------------- ----(((((((((....)).))....((((((....((((.((((....)))).))))......))))))..........((((......))))...)))))...------------- ( -28.70, z-score = -2.99, R) >droEre2.scaffold_4690 18357285 101 - 18748788 ----UUCCAGCUCCUCGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAGAAAUUAUCAGCGCACACAGCAGUGCAAAGGGAAAAA------------- ----...(((((....)))))((......(((((..((((.((((....)))).))))((.....))........)))))((((......))))...))......------------- ( -29.60, z-score = -1.51, R) >droAna3.scaffold_12929 2017091 98 + 3277472 ---AUUCGCAACUCUCGGCUGCCAUAUUUGCCAAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGG-GAAAAUUAUCAACGCACACGGAAAAGUGCGGGUAA---------------- ---.........(((((((..........)))....((((.((((....)))).))))......))-))...(((((..(((((........))))))))))---------------- ( -23.90, z-score = -0.89, R) >dp4.chrXL_group1a 702209 107 + 9151740 -----------CGGGUGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCGAAAAUUAUCAACGCACACAGAGAAGUGCAGCAGGCAGGGUACUGGGACGA -----------(.((((.(((((...((((((....((((.((((....)))).))))......))))))..........((((........))))....)))).).)))).)..... ( -32.30, z-score = -1.70, R) >droPer1.super_15 1338968 107 + 2181545 -----------GGGGUGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCGAAAAUUAUCAACGCACACAGAGAAGUGCAGCAGGCAGGGUACUGGGACGA -----------..((((.(((((...((((((....((((.((((....)))).))))......))))))..........((((........))))....)))).).))))....... ( -30.60, z-score = -1.17, R) >droGri2.scaffold_15203 1049598 105 + 11997470 CAACUGUCUACUGUCUAUGUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCACAUUAAGCACAAAUUAUCAACGUACACAGUGGCACAGGGGCUAAGA------------- (..(((((((((((.(((((.....(((((((.((.((((.((((....)))).))))...)).))))..))).....))))).))))))).))))..)......------------- ( -30.90, z-score = -2.28, R) >droMoj3.scaffold_6473 7259675 83 - 16943266 ----CAACUGCCGUCUGUCUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCGCAUUAAGCAGAAAUUAUCAACGUACACA------------------------------- ----.............(((((......(((.....((((.((((....)))).)))))))....))))).................------------------------------- ( -16.60, z-score = -0.31, R) >droVir3.scaffold_12970 4290219 99 - 11907090 ----CAACUGCUGUCUGUCUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCGCAUUAAGCAAAAAUUAUCAACGCACACAAAGAGAGCAAGAGAAA--------------- ----....((((.(((((.(((.(((((((((.((.((((.((((....)))).))))...)).))))))...)))....))).))..))).)))).......--------------- ( -24.20, z-score = -1.16, R) >consensus _______CCGCCGUCCGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAAAAAUUAUCAACGCACACAGAAGAGCAAAGGAAAAAA_____________ ..........................((((((....((((.((((....)))).))))......))))))................................................ (-13.43 = -13.08 + -0.35)



| Location | 22,009,647 – 22,009,751 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 65.11 |
| Shannon entropy | 0.70303 |
| G+C content | 0.46003 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 22009647 104 - 22422827 --------------CCGGGUAGGUUAUCCCGCCCUCUUGCCCUGCUCCGCCAUCCAGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGGCGCAUUAGGCAAAAAUUAUCAA --------------..(((..((....))..)))..(((((......((((...((((..........))))........((((....))))..)))).....))))).......... ( -28.60, z-score = -0.71, R) >droSec1.super_55 162879 104 - 187931 --------------CCGGGUAGGUUAUCCCACCCUCUUGCCCUGCUCCGCCAUCCAGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAAAAAUUAUCAA --------------..((((.((....)).))))..(((((.(((.........((((..........))))...((((.((((....)))).)))))))...))))).......... ( -29.90, z-score = -2.46, R) >droEre2.scaffold_4690 18357311 87 - 18748788 ------------------------------GUGGCUCCUGCUUU-CCAGCUCCUCGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAGAAAUUAUCAG ------------------------------(((((.((.(((..-..))).....))..))))).((((((....((((.((((....)))).))))......))))))......... ( -23.00, z-score = -0.57, R) >droAna3.scaffold_12929 2017114 111 + 3277472 ----UAGUCCUGAGUCCUUCACUCGGGUAGGUGCUUAUCAAUUC-GCAACUCUCGGC-UGCCAUAUUUGCCAAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGGA-AAAUUAUCAA ----....(((((((.....)))))))..(((((..........-.........(((-..........)))....((((.((((....)))).))))))))).....-.......... ( -25.60, z-score = -0.43, R) >dp4.chrXL_group1a 702248 92 + 9151740 ----------------------CCGAGAGUGGGUAGGCGGGCUG----GUCGGGUGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCGAAAAUUAUCAA ----------------------.........(((((((((.(..----.......).)))))...((((((....((((.((((....)))).))))......))))))...)))).. ( -25.30, z-score = -0.24, R) >droPer1.super_15 1339007 83 + 2181545 -------------------------------CCAUGGCGGGCUG----GUGGGGUGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCGAAAAUUAUCAA -------------------------------..(((((((.(..----.......).))))))).((((((....((((.((((....)))).))))......))))))......... ( -25.30, z-score = -1.24, R) >droGri2.scaffold_15203 1049624 115 + 11997470 CGAGCAAACUUCUGUCUUUCAGGCAA--AGGUGCUUGUCAACUG-UCUACUGUCUAUGUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCACAUUAAGCACAAAUUAUCAA .(((((((....((((.....)))).--.((..(..(.((....-.....)).)...)..))...)))))))...((((.((((....)))).))))..................... ( -28.20, z-score = -1.03, R) >droMoj3.scaffold_6473 7259683 111 - 16943266 CUGGCAAACUUCUGUCUCUCCGGCAA--AGGUGCUUGUCAACUG-----CCGUCUGUCUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCGCAUUAAGCAGAAAUUAUCAA .(((((.((...((((.....)))).--.(((...........)-----))....)).)))))..((((((.((.((((.((((....)))).))))...)).))))))......... ( -27.40, z-score = -0.56, R) >droVir3.scaffold_12970 4290243 111 - 11907090 CUGGCAAACUUCUGUCUCUCAGGCAA--AGGUGCUUGUCAACUG-----CUGUCUGUCUGCCAUAUUUGCUCAAAGACUGAAGUGCAAACUUGAGUCGCAUUAAGCAAAAAUUAUCAA .(((((.((....))....((((((.--.(((........))).-----.))))))..)))))..((((((.((.((((.((((....)))).))))...)).))))))......... ( -29.90, z-score = -1.56, R) >consensus ______________UC__UCAGGCAA__AGGUGCUUGUCAACUG__C_GCCGUCUGGCUGCCAUAUUUGCUGAAAGACUAAAGUGCAAACUUGAGUCGCAUUAGGCAAAAAUUAUCAA .................................................................((((((....((((.((((....)))).))))......))))))......... (-12.90 = -12.73 + -0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:31 2011