| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,902,724 – 21,902,795 |
| Length | 71 |
| Max. P | 0.966004 |

| Location | 21,902,724 – 21,902,795 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.54662 |
| G+C content | 0.40740 |
| Mean single sequence MFE | -9.70 |
| Consensus MFE | -5.89 |
| Energy contribution | -5.61 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
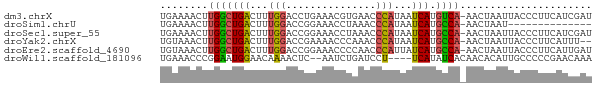
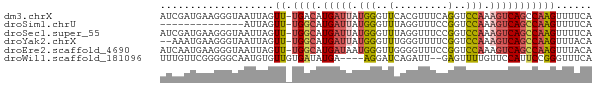
>dm3.chrX 21902724 71 + 22422827 UGAAAACUUGGCUGACUUUGGACCUGAAACGUGAACCCAUAAUCAUGUCA-AACUAAUUACCCUUCAUCGAU .......((((((((...(((...............)))...))).))))-).................... ( -7.46, z-score = 0.20, R) >droSim1.chrU 2241528 57 - 15797150 UGAAAACUUGGCUGACUUUGGACCGGAAACCUAAACCCAUAAUCAUGCCA-AACUAAU-------------- .......((((((((...(((...(....)......)))...))).))))-)......-------------- ( -11.50, z-score = -3.44, R) >droSec1.super_55 62638 71 + 187931 UGAAAACUUGGCUGACUUUGGACCGGAAACCUAAACCCAUAAUCAUGCCA-AACUAAUUACCCUUCAUCGAU .......((((((((...(((...(....)......)))...))).))))-).................... ( -11.50, z-score = -2.25, R) >droYak2.chrX 21346119 69 + 21770863 UGUAAACUUGGCUGACUUUGGACCGAAAACCCAAACCCAUAAUCAUGCCA-AACUAAUUACCCUUCAUUU-- .......((((((((...(((...............)))...))).))))-)..................-- ( -10.16, z-score = -2.22, R) >droEre2.scaffold_4690 18278982 71 + 18748788 UGUAAACUUGGCUGACUUUGGACCGGAAACCCCAACCCAUUAUCAUGCCA-AACUAAUUACCCUUCAUUGAU .......((((((((...(((...((.....))...)))...))).))))-).................... ( -12.80, z-score = -1.99, R) >droWil1.scaffold_181096 7234666 66 + 12416693 UGAAACCCGGAAUGGAACAAAACUC--AAUCUGAUCCU----UCAUAUCACAACACAUUGCCCCCGAACAAA ((((...((((.(((........))--).))))....)----)))........................... ( -4.80, z-score = 0.68, R) >consensus UGAAAACUUGGCUGACUUUGGACCGGAAACCUAAACCCAUAAUCAUGCCA_AACUAAUUACCCUUCAUCGAU .........((((((...(((...............)))...))).)))....................... ( -5.89 = -5.61 + -0.28)
| Location | 21,902,724 – 21,902,795 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.54662 |
| G+C content | 0.40740 |
| Mean single sequence MFE | -15.72 |
| Consensus MFE | -8.26 |
| Energy contribution | -8.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
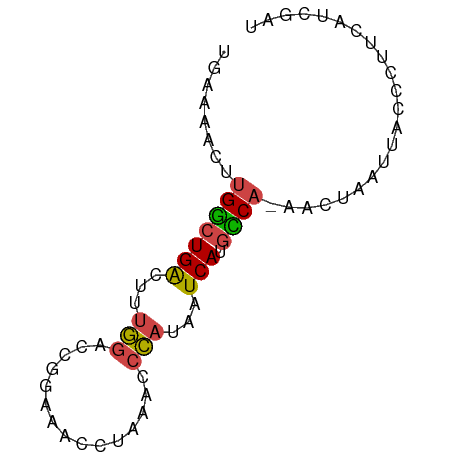
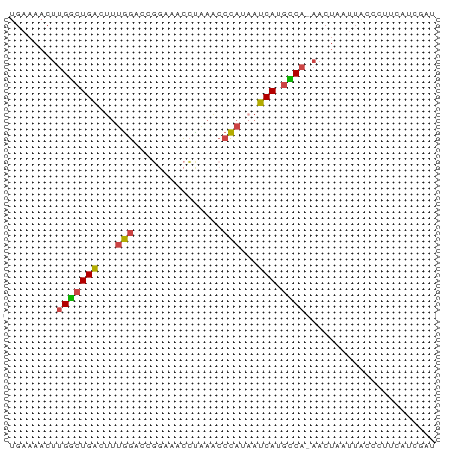
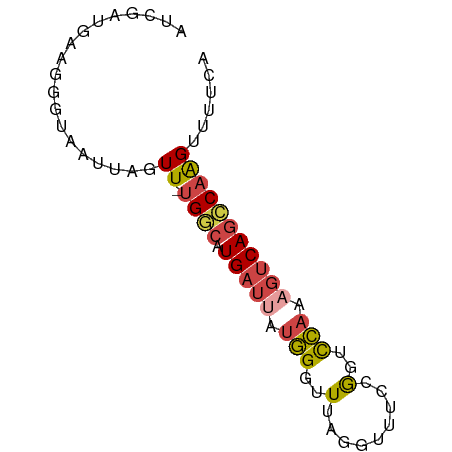
>dm3.chrX 21902724 71 - 22422827 AUCGAUGAAGGGUAAUUAGUU-UGACAUGAUUAUGGGUUCACGUUUCAGGUCCAAAGUCAGCCAAGUUUUCA ...((.((..(((......((-.(((.(((..(((......))).))).))).)).....)))...)).)). ( -10.50, z-score = 0.69, R) >droSim1.chrU 2241528 57 + 15797150 --------------AUUAGUU-UGGCAUGAUUAUGGGUUUAGGUUUCCGGUCCAAAGUCAGCCAAGUUUUCA --------------.....((-((((.(((((.((((....((...))..)))).)))))))))))...... ( -15.60, z-score = -3.01, R) >droSec1.super_55 62638 71 - 187931 AUCGAUGAAGGGUAAUUAGUU-UGGCAUGAUUAUGGGUUUAGGUUUCCGGUCCAAAGUCAGCCAAGUUUUCA .....((((((........((-((((.(((((.((((....((...))..)))).))))))))))))))))) ( -15.90, z-score = -1.13, R) >droYak2.chrX 21346119 69 - 21770863 --AAAUGAAGGGUAAUUAGUU-UGGCAUGAUUAUGGGUUUGGGUUUUCGGUCCAAAGUCAGCCAAGUUUACA --.........((((....((-((((.(((((.(((..(.........)..))).))))))))))).)))). ( -16.70, z-score = -1.91, R) >droEre2.scaffold_4690 18278982 71 - 18748788 AUCAAUGAAGGGUAAUUAGUU-UGGCAUGAUAAUGGGUUGGGGUUUCCGGUCCAAAGUCAGCCAAGUUUACA ...........((((....((-((((.((((..(((..(((.....)))..)))..)))))))))).)))). ( -21.60, z-score = -2.77, R) >droWil1.scaffold_181096 7234666 66 - 12416693 UUUGUUCGGGGGCAAUGUGUUGUGAUAUGA----AGGAUCAGAUU--GAGUUUUGUUCCAUUCCGGGUUUCA ...(..(((((((((....)))).......----.(((.((((..--....)))).))).)))))..).... ( -14.00, z-score = -0.04, R) >consensus AUCGAUGAAGGGUAAUUAGUU_UGGCAUGAUUAUGGGUUUAGGUUUCCGGUCCAAAGUCAGCCAAGUUUUCA ......................((((.(((((.(((..(.........)..))).)))))))))........ ( -8.26 = -8.82 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:27 2011