
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,859,884 – 21,859,982 |
| Length | 98 |
| Max. P | 0.601078 |

| Location | 21,859,884 – 21,859,982 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.50327 |
| G+C content | 0.63417 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21859884 98 + 22422827 ACUAUCCCACCUCUUACUCAACGUUCUCCGCCAACAGCG-GCUCCUCAAGCGCCUCGUAUGCGCCGCGCUCCAGUGUGCAGAGGAGCAGCGUUGGUACC--- ..............(((.(((((((...(((.....)))-(((((((..((((.......)))).((((......)))).)))))))))))))))))..--- ( -37.70, z-score = -3.64, R) >droEre2.scaffold_4690 18238533 98 + 18748788 ACUAUCCCAGCUCGUACUCGACGUACUCCGCCAACAGCG-GCUCCUCCAGCUCCUCGUAUGCGCCGCGCUCCAGCGUUCAGAGGUGCAGCGUUGGUUCC--- ......(((((.(((((.....)))).((((.....)))-)........((.((((....((((.(.....).))))...)))).)).).)))))....--- ( -29.10, z-score = -0.46, R) >droAna3.scaffold_13117 3833400 101 - 5790199 ACUACCCCAGCUCCUAUUCAACGUACUCCUCCAGCCACA-CCAGCGGCUCCUCGUCGUACACGCCCCGCUCCAGCCUGCAGAGGAGCAGCGUCGGGUCCAGC ...((((...............((((..(...((((.(.-...).))))....)..))))((((...(((((..........))))).)))).))))..... ( -25.70, z-score = -0.26, R) >droYak2.chrX 21305497 98 + 21770863 ACUAUCCCAGCUCGUACUCGACAUACUCAGCCAACAGCG-GCUCCUCCAGCUCCUCGUAUGCGCCGCGCUCCAGCGUGCAGAGGAGCAGCGUUGGUUCC--- .........(.(((....))))......(((((((.((.-(((((((..((.........))(((((......))).)).))))))).)))))))))..--- ( -34.60, z-score = -1.91, R) >droSec1.super_55 15918 98 + 187931 ACUAUCCCAGCUCGUACUCAACGUACUCCGCCAACAGCG-GCUCCUCAAGCGCCUCUUAUGCGCCGCGCUCCAGUGUGCAGAGGAGCAGCGUUGGUACC--- .............((((.((((((....(((.....)))-(((((((..((((.......)))).((((......)))).))))))).)))))))))).--- ( -40.80, z-score = -3.92, R) >droSim1.chrX_random 5662782 98 + 5698898 ACUAUCCCAGCUCGUACUCAACGUACUCCGCCAACAGCG-GCUCCUCAAGCGCCUCUUAUGCGCCGCGCUCCAGUGUGCAGAGGAGCAGCGUUGGUACC--- .............((((.((((((....(((.....)))-(((((((..((((.......)))).((((......)))).))))))).)))))))))).--- ( -40.80, z-score = -3.92, R) >anoGam1.chr3R 10778820 98 + 53272125 ACUUUCUCGGCGGUGUAACAGCG-GCAGCGGCAACACAGUACGGCGCCGGUUCCUCGCUUGCCGGGACGGCCGGUGGACAGGCGCAGGGCGUCACCAGC--- ........((((.((((...((.-...))(....)....)))).))))(((((((((((((((((.....))))....)))))).))))....)))...--- ( -36.30, z-score = 1.34, R) >consensus ACUAUCCCAGCUCGUACUCAACGUACUCCGCCAACAGCG_GCUCCUCCAGCUCCUCGUAUGCGCCGCGCUCCAGUGUGCAGAGGAGCAGCGUUGGUACC___ .............................((((((.....(((.....)))..............(((((((..........))))).))))))))...... (-13.86 = -13.87 + 0.01)

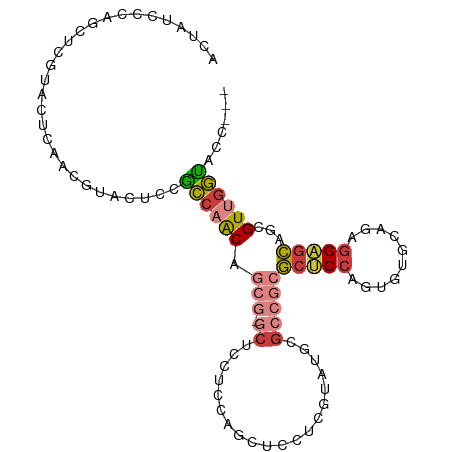

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:25 2011