| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,799,876 – 21,800,005 |
| Length | 129 |
| Max. P | 0.527610 |

| Location | 21,799,876 – 21,800,005 |
|---|---|
| Length | 129 |
| Sequences | 4 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 61.58 |
| Shannon entropy | 0.63263 |
| G+C content | 0.56713 |
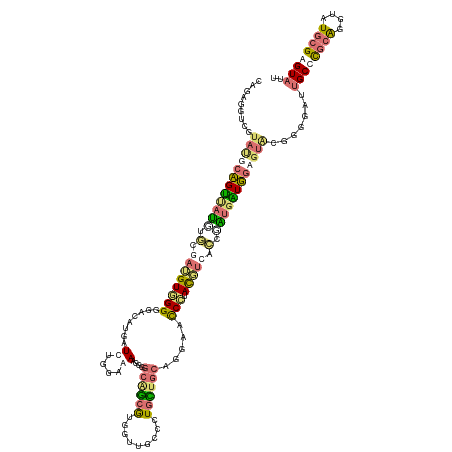
| Mean single sequence MFE | -49.25 |
| Consensus MFE | -20.26 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.66 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
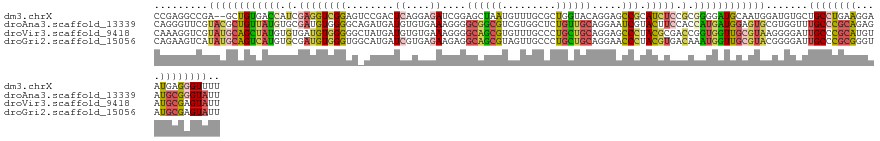
>dm3.chrX 21799876 129 + 22422827 CCGAGGCCGA--GCUGUGACCAUCGAGGUCGGAGUCCGACUCAGGAGAUCGGAGCUAAUGUUUGCGCUGGUACAGGAGCCGCAUCUCCGCGGGGAUGCAAUGGAUGUGCUGCCUGAAGGAAUGAGGGUUUU ..(((((((.--.((.(((((.....))))).))..)..((((.....((((.((........))((.((((((....(((((((((....)))))))...)).)))))))))))).....)))))))))) ( -47.00, z-score = -0.16, R) >droAna3.scaffold_13339 3493958 131 - 4599533 CAGGGUUCGUACGCUGUUAUGUGCGAUGUGGGGCAGAUGAUGUGUGAAAGGGGCGGCGUCGUGGCUCUGUUGCAGGAAUCGUACUUCCACCAUGAUGGAGUGCGUGGUUUGCCCGCAGAGAUGCGGGUAUU .......(((((.((((((((((((((..(((((..(((((((((.......)).))))))).)))))))))))((((......))))..)))))))).))))).....((((((((....)))))))).. ( -49.90, z-score = -1.48, R) >droVir3.scaffold_9418 1350 131 - 1532 CAAAGGUCGUAUGCAGCUAUGUGUGAUGUGGGGGCUAUGAUGUGUGAAAGGGGCAGCGUGUUUGCCCUGCUGCAGGAGCCCUACGCGACCGGUGGUUGCGUAAGGGGAUUGCCCGCAUGUAUGCGAGUAUU ....(.((((((((.((.....))...(((.((((.....(.((((...(((((((.....)))))))..)))).)..((((((((((((...)))))))).))))....)))).))))))))))).)... ( -52.80, z-score = -1.75, R) >droGri2.scaffold_15056 31386 131 - 38174 CAGAAGUCAUAUGCAGUCAUGUGCGAUGUGGGUGGCAUGAUCGUGAGAAGAGGCAGCGUAGUUGCCCUGCUGCAGGAACCCUACGUGACAAAUGGUUGCGUACGGGGAUUGCCCGCGGGUAUGCGAGUAUU ....(.(((((((((......))).)))))).).(((((..((((((.((.((((((...)))))))).))((((...(((((((..((.....))..)))).)))..)))).))))..)))))....... ( -47.30, z-score = -1.05, R) >consensus CAGAGGUCGUAUGCAGUUAUGUGCGAUGUGGGGGACAUGAUCUGGAAAAGGGGCAGCGUGGUUGCCCUGCUGCAGGAACCCUACGUCACCGAUGAUGGAGUACGGGGAUUGCCCGCAGGUAUGCGAGUAUU ......((....(((.(((((.....))))).))).................((((((.........)))))).))..(((((((((((.....))))))))..)))..((((((((....)))))))).. (-20.26 = -19.95 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:20 2011