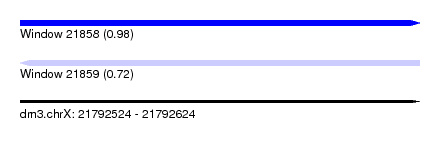
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,792,524 – 21,792,624 |
| Length | 100 |
| Max. P | 0.977186 |

| Location | 21,792,524 – 21,792,624 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 63.42 |
| Shannon entropy | 0.65246 |
| G+C content | 0.50513 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -16.60 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
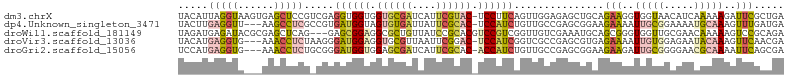
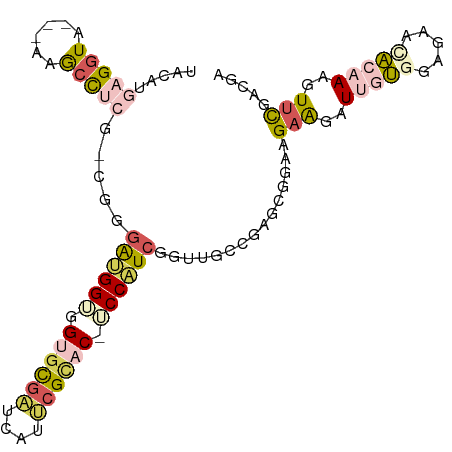
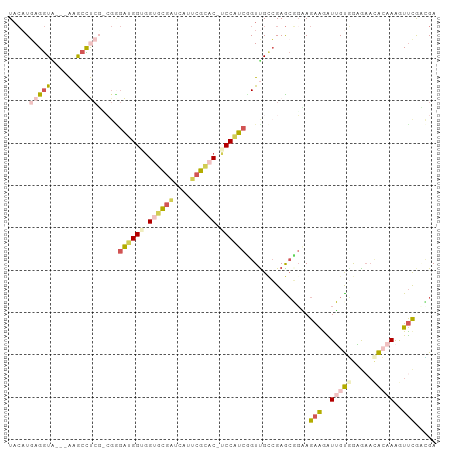
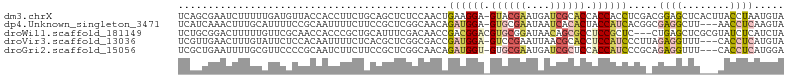
>dm3.chrX 21792524 100 + 22422827 UACAUUAGGUAAGUGAGCUCCGUCGAGGUGGUGGUGCGAUCAUUCGUAC-UCCUUCAGUUGGAGAGCUGCAGAAGGUGGUAACAUCAAAAAGAUUCGCUGA ...........(((((((((..((.((.(((.(((((((....))))))-)...))).)).)))))).......((((....))))........))))).. ( -27.20, z-score = -0.32, R) >dp4.Unknown_singleton_3471 1043 97 + 15135 UACUUGAGGUU---AAGCCUCGCCGUGAUGGUAGUGUGAUUAUUCGCAC-UCCAUCUGUUGCCGAGCGGAAGAAAAUUGCGGAAAAUGCAAAGUUUGAUGA .((((((((..---...)))).(((..((((.(((((((....))))))-))).((((((....)))))).....))..)))........))))....... ( -31.40, z-score = -2.74, R) >droWil1.scaffold_181149 395062 98 - 589524 UAGAUGAGAUACGCGAGCUCAG---GAGCGGAGGCGCUGUUAUCCGCACGUCCGUCGGUUGUCGAAAUGCAGCGGGUGGUUGCGAACAAAAAGUCCGCAGA ....((((...(....))))).---..(((((.....((((...((((...((..(.(((((......))))).)..)).)))))))).....)))))... ( -28.70, z-score = 1.06, R) >droVir3.scaffold_13036 417552 97 + 1135500 UACAUGAGGUG---AAACCUCUAAGGGAUGGAGGUGCGUUAAUUCGGAC-UCCAUCGGUCGCCGAGCGUGAGAAAAUUGUGGAGAAUACAAAGUUCAACGA ..((((.((((---(..((.....))(((((((.(.((......)).))-))))))..)))))...)))).(((..(((((.....)))))..)))..... ( -30.90, z-score = -2.36, R) >droGri2.scaffold_15056 30727 97 - 38174 UCCAUGAGGUG---AAACCUCUGCGGGAUGGUGGAGCGAUCAUUCGCAC-ACCAUCUGUUGCCGAGCGGAAGAAGAUUGCGGGGAACGCAAAAUUCAGCGA (((..((((..---...)))).(((((((((((..((((....)))).)-)))))))..))).....))).(((..(((((.....)))))..)))..... ( -38.00, z-score = -2.62, R) >consensus UACAUGAGGUA___AAGCCUCG_CGGGAUGGUGGUGCGAUCAUUCGCAC_UCCAUCGGUUGCCGAGCGGAAGAAGAUUGUGGAGAACACAAAGUUCGACGA .....(((((......))))).....((((((.((((((....)))))).))))))...............(((..(((((.....)))))..)))..... (-16.60 = -18.28 + 1.68)
| Location | 21,792,524 – 21,792,624 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 63.42 |
| Shannon entropy | 0.65246 |
| G+C content | 0.50513 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -9.60 |
| Energy contribution | -11.28 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21792524 100 - 22422827 UCAGCGAAUCUUUUUGAUGUUACCACCUUCUGCAGCUCUCCAACUGAAGGA-GUACGAAUGAUCGCACCACCACCUCGACGGAGCUCACUUACCUAAUGUA ...(((((((.....)))........(((((.(((........))).))))-).........))))........(((....)))................. ( -14.20, z-score = 1.27, R) >dp4.Unknown_singleton_3471 1043 97 - 15135 UCAUCAAACUUUGCAUUUUCCGCAAUUUUCUUCCGCUCGGCAACAGAUGGA-GUGCGAAUAAUCACACUACCAUCACGGCGAGGCUU---AACCUCAAGUA ..........((((.......)))).........(((.(....).((((((-(((.((....)).)))).)))))..)))((((...---..))))..... ( -25.00, z-score = -2.83, R) >droWil1.scaffold_181149 395062 98 + 589524 UCUGCGGACUUUUUGUUCGCAACCACCCGCUGCAUUUCGACAACCGACGGACGUGCGGAUAACAGCGCCUCCGCUC---CUGAGCUCGCGUAUCUCAUCUA ..(((((((.....)))))))......(((.((...(((.....)))((((.((((........)))).))))...---....))..)))........... ( -28.10, z-score = -1.06, R) >droVir3.scaffold_13036 417552 97 - 1135500 UCGUUGAACUUUGUAUUCUCCACAAUUUUCUCACGCUCGGCGACCGAUGGA-GUCCGAAUUAACGCACCUCCAUCCCUUAGAGGUUU---CACCUCAUGUA (((((((...((((.......))))...........)))))))..((((((-(..((......))...))))))).....((((...---..))))..... ( -20.94, z-score = -1.14, R) >droGri2.scaffold_15056 30727 97 + 38174 UCGCUGAAUUUUGCGUUCCCCGCAAUCUUCUUCCGCUCGGCAACAGAUGGU-GUGCGAAUGAUCGCUCCACCAUCCCGCAGAGGUUU---CACCUCAUGGA .....(((..(((((.....)))))..))).(((((..(....).((((((-(.((((....))))..)))))))..)).((((...---..))))..))) ( -34.80, z-score = -2.99, R) >consensus UCAGCGAACUUUGUAUUCGCCACAAUCUUCUGCCGCUCGGCAACAGAUGGA_GUGCGAAUAAUCGCACCACCAUCCCG_CGAGGCUU___CACCUCAUGUA .............................................((((((.((((((....)))))).)))))).....((((........))))..... ( -9.60 = -11.28 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:17 2011