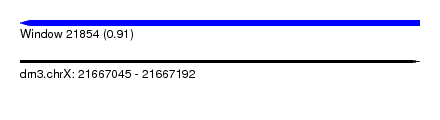
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,667,045 – 21,667,192 |
| Length | 147 |
| Max. P | 0.913727 |

| Location | 21,667,045 – 21,667,192 |
|---|---|
| Length | 147 |
| Sequences | 4 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Shannon entropy | 0.35389 |
| G+C content | 0.38342 |
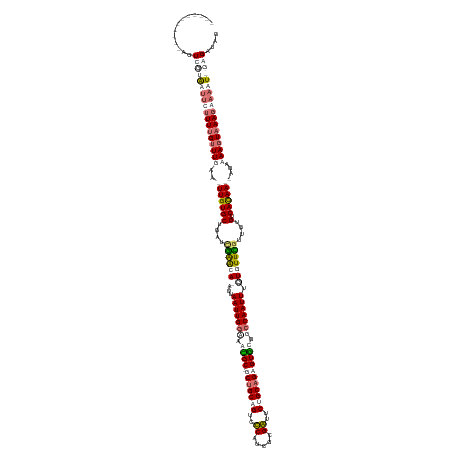
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -24.10 |
| Energy contribution | -26.35 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21667045 147 - 22422827 AGUCAGAAAUUUUUUCGAUCCCUUUUGUUUUAA-UUGUGCUGUUUGGACAUAGUAAUUGGCAGUGC-UCUGCCGUCCCAUCGCGGUUCUGCAGAGUACGGCCAAUUUGUUUUUGUUGUGUCACAA--AGAAAAGUAAAUAAAAU-GAGAUAG .(((.(((.....))))))...(((..(((((.-...((((.(((((((((((.(((((((.((((-(((((.(..((.....))..).))))))))).)))))))........))))))).)))--)....))))..))))).-.)))... ( -43.20, z-score = -3.51, R) >droSim1.chrU 13094527 140 + 15797150 -----------AGUCGUAAUUUUUUUGUUUGGAUUUGUGCUGAUCGAGUA-AUUAAUUGGAAACGCGGCUGCAGUCUCAUCGCGAUUCUGCAGAGUGCGGCCAAUUUGUGUUCGUUGUGUCAUAAAGAGAAAAGUAAAGAAAAUAGAGAUAG -----------.(((...((((((((.(((...(((((((....((((((-...((((((...((((.((((((..((.....))..))))))..)))).))))))..))))))....).))))))..))).....))))))))...))).. ( -37.70, z-score = -1.49, R) >droSec1.super_49 60662 140 + 245908 -----------AGUCGUAAUUUUUUUGUUUGGAUUUGUGCUGAUCGAGUA-AUUAAUUGGAAACGCGGCUGCAGUCUCAUCGCGAUUCUGCAGAGUGCGGACAAUUUGUGUUCGUUGUGUCAUAAAGAGAAAAGUAAAGAAAAUAGAGAUAG -----------.(((...((((((((.(((...(((((((....((((((-...(((((....((((.((((((..((.....))..))))))..))))..)))))..))))))....).))))))..))).....))))))))...))).. ( -34.10, z-score = -0.44, R) >droEre2.scaffold_4690 18049892 147 - 18748788 AGUCAGAAGAUAUUUUUGUUUCUUUCGUUUUAA-UUGUGCUGUUCAGACAUAUUAAUUGGUAGUGC-UCUGCCGUCCCAUCGCGGUUCUGCUGAGUGCGGCCAAUUUAUUUUUAAUGUGUCACAA--AGAAAACUAAAGAAAAU-GAGAUAG ..........((((((..(((((((.(((((..-((((........(((((((((((((((.(..(-((.((.(..((.....))..).)).)))..).)))))).......)))))))))))))--..))))).)))))))..-)))))). ( -41.41, z-score = -3.21, R) >consensus ___________AGUCGUAAUUCUUUUGUUUGAA_UUGUGCUGAUCGAACA_AUUAAUUGGAAACGC_GCUGCAGUCCCAUCGCGAUUCUGCAGAGUGCGGCCAAUUUGUGUUCGUUGUGUCACAA__AGAAAAGUAAAGAAAAU_GAGAUAG .............((....((((((..((((...((((((....(((((((...(((((((.((((.(((((((..((.....))..))))))))))).))))))).)))))))....).)))))...))))...))))))....))..... (-24.10 = -26.35 + 2.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:13 2011