| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,664,898 – 21,665,015 |
| Length | 117 |
| Max. P | 0.507371 |

| Location | 21,664,898 – 21,665,015 |
|---|---|
| Length | 117 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Shannon entropy | 0.45542 |
| G+C content | 0.51068 |
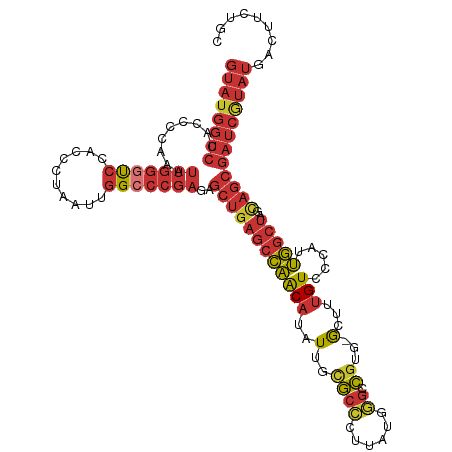
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -22.13 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
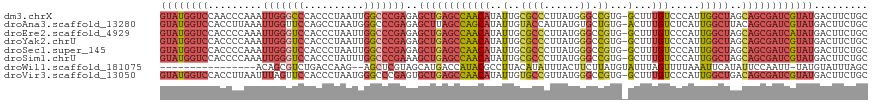
>dm3.chrX 21664898 117 - 22422827 GUAUGGUCCAACCCAAAUUGGGCCCACCCUAAUUGGCCCGAGAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUAGCAGCGAUCGUAUGACUUCUGC ((((((((.........(((((((..........)))))))..((((((((((((..(..(((((....))).))..-)...))).....)))))..))))))))))))......... ( -40.70, z-score = -1.46, R) >droAna3.scaffold_13280 125880 117 + 1428709 GUAUGGUCCACCUUAAAUUGGUUCCAGCCUAAUGGGCCCGAGAGCUUAGCCAGCAUAUUGUACCAUUAUGUGCUGUG-ACUUUGUCUCAUUGGCUUACAGCGAUCGUAUGACUUCUGC ((((((((.........((((..(((......)))..))))..(((.(((((((((((.........))))))((.(-((...))).)).)))))...)))))))))))......... ( -33.80, z-score = -1.29, R) >droEre2.scaffold_4929 24691821 117 - 26641161 GUAUGGUCCACCCCAAAUUGGGUCCACCCUAAUUGGCCCGAGAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUAGCAGCGAUCAUAUGACUUCUGC ((((((((.........(((((.(((.......))))))))..((((((((((((..(..(((((....))).))..-)...))).....)))))..))))))))))))......... ( -39.40, z-score = -1.25, R) >droYak2.chrU 21630169 117 + 28119190 GUAUGGUCCACCCCAAAUUGGGUCCACCCUAAUUGGCCCGAGAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUAGCAGCGAUCGUAUGACUUCUGC ((((((((.........(((((.(((.......))))))))..((((((((((((..(..(((((....))).))..-)...))).....)))))..))))))))))))......... ( -39.00, z-score = -1.02, R) >droSec1.super_145 9788 117 - 49412 GUAUGGUCCACCCCAAAUUGGGUCCACCCUAAUUGGCCCGAGAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUAGCAGCGAUCGUAUGACUUCUGC ((((((((.........(((((.(((.......))))))))..((((((((((((..(..(((((....))).))..-)...))).....)))))..))))))))))))......... ( -39.00, z-score = -1.02, R) >droSim1.chrU 2224577 117 - 15797150 GUAUGGUCCACCCCAAAUUGGGUCCACCCUAUUUGGCCCGAAAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUAGCAGCGAUCGUAUGACUUCUGC ((((((((....((((((.(((....))).)))))).......((((((((((((..(..(((((....))).))..-)...))).....)))))..))))))))))))......... ( -40.60, z-score = -1.78, R) >droWil1.scaffold_181075 769905 99 + 1141854 ----------------ACAGCGUCUGACCAAG--AGCUCGUAGCAUGACCAUACGCCUUACAUAUUUACUUCUUAUGUAUUUAGUUUUAAAUUCAUAUUCCAAUU-UAUGUAUUUAGC ----------------...((((.((..((..--.((.....)).))..)).))))..((((((.........)))))).......((((((.((((........-)))).)))))). ( -13.60, z-score = -0.54, R) >droVir3.scaffold_13050 3111612 117 + 3426691 GUAUGGUCCACCUUAAUUUAGUUCCACCCUAAUGGGCCCGAGUGCUGAGCCAACAUAUUGUGCCGUUAUGGGCCGUG-GCUUUGUCCCAUUGGCUGACAGCGAUCGUAUGACUUCUGC ((((((((.......(((..(..(((......)))..)..)))(((((((((.......(.(((......))))(((-(.......)))))))))..))))))))))))......... ( -30.90, z-score = 0.63, R) >consensus GUAUGGUCCACCCCAAAUUGGGUCCACCCUAAUUGGCCCGAGAGCUGAGCCAACAUAUUGCGCCCUUAUGGGCCGUG_GCUUUGUCCCAUUGGCUAGCAGCGAUCGUAUGACUUCUGC ((((((((.........(((((((..........)))))))..((((((((((.(((..(((((......))).....))..)))....))))))..))))))))))))......... (-22.13 = -23.65 + 1.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:12 2011