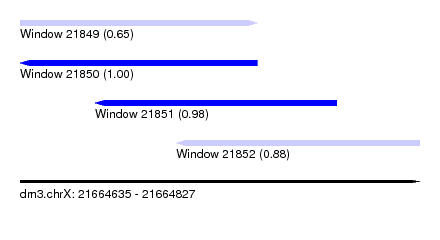
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,664,635 – 21,664,827 |
| Length | 192 |
| Max. P | 0.995007 |

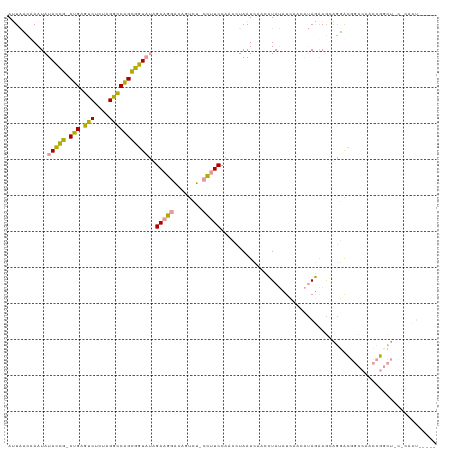
| Location | 21,664,635 – 21,664,749 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 56.09 |
| Shannon entropy | 0.61723 |
| G+C content | 0.45684 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -16.99 |
| Energy contribution | -15.57 |
| Covariance contribution | -1.42 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21664635 114 + 22422827 ----CAGUGGGGAGCCGGUUGGCCGUCUUGUUUCUAUGAUGAAAUAGGAUGAUUACGUUGGAAGG-AGACUGUCCUUCAAUACCGCUGGCCUAGAGACUCAGCCGGUAUAUUUUGUAAU ----.........(((((((((((((((((((((......))))))))))......((..(((((-(.....))))))...))....))))..(.....))))))))............ ( -35.70, z-score = -1.38, R) >droWil1.scaffold_181075 769677 103 - 1141854 ------------CGGUGGUGGG--AUCCAACUUAUGGGUAGAUUUAUGUUGAUUAGGA-GGAGAUACGAUUAAGUAUCUGAAUCACCGUCCUUGUGGAUCAG-CAAUAUAUUAGAAAUU ------------...(((((..--(((((.....))))).......((((((((((((-((((((((......))))))......)).))))....))))))-))...)))))...... ( -22.50, z-score = -0.02, R) >droVir3.scaffold_13050 3111345 118 - 3426691 CCAAUAGUGCGUAGCCGGUUGGCCGCACUGCUCCUAUGUUGAGAUAGGGAGAUUAGGGUGGAAGGGAAACUGUCCUUCGAUACCGCUGGCC-AUAGACUCAGUCGGUAUAUUGGUUAAU (((((((((((..(((....))))))))).(((((.(((....))))))))....(((.(((.((....)).)))))).((((((((((..-......)))).)))))))))))..... ( -39.20, z-score = -0.91, R) >consensus _____AGUG_G_AGCCGGUUGGCCGUCCUGCUUCUAUGUUGAAAUAGGAUGAUUAGGAUGGAAGG_AGACUGUCCUUCAAUACCGCUGGCCUAGAGACUCAG_CGGUAUAUUAGAUAAU .............((((((.((.(((((((.((((((......))))))....)))))))(((((........)))))....))))))))............................. (-16.99 = -15.57 + -1.42)

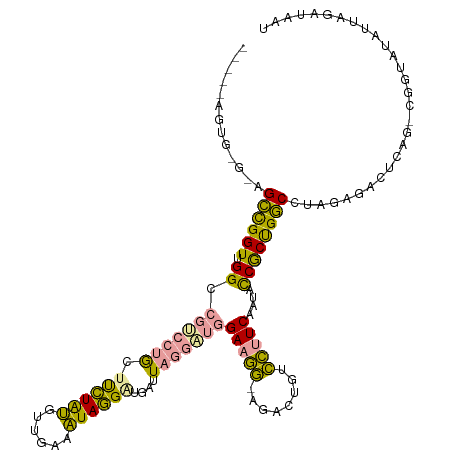
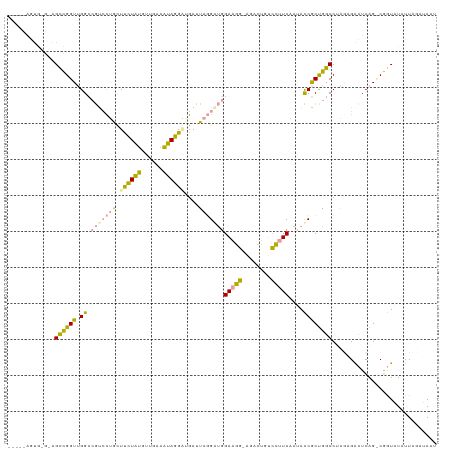
| Location | 21,664,635 – 21,664,749 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 56.09 |
| Shannon entropy | 0.61723 |
| G+C content | 0.45684 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -13.58 |
| Energy contribution | -12.37 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21664635 114 - 22422827 AUUACAAAAUAUACCGGCUGAGUCUCUAGGCCAGCGGUAUUGAAGGACAGUCU-CCUUCCAACGUAAUCAUCCUAUUUCAUCAUAGAAACAAGACGGCCAACCGGCUCCCCACUG---- (((((.....((((((.(((.(((....)))))))))))).((((((.....)-)))))....)))))....((((......)))).........((((....))))........---- ( -27.70, z-score = -1.86, R) >droWil1.scaffold_181075 769677 103 + 1141854 AAUUUCUAAUAUAUUG-CUGAUCCACAAGGACGGUGAUUCAGAUACUUAAUCGUAUCUCC-UCCUAAUCAACAUAAAUCUACCCAUAAGUUGGAU--CCCACCACCG------------ ....((((((..((..-(((.(((....))))))..))..((((((......))))))..-...........................)))))).--..........------------ ( -17.30, z-score = -1.46, R) >droVir3.scaffold_13050 3111345 118 + 3426691 AUUAACCAAUAUACCGACUGAGUCUAU-GGCCAGCGGUAUCGAAGGACAGUUUCCCUUCCACCCUAAUCUCCCUAUCUCAACAUAGGAGCAGUGCGGCCAACCGGCUACGCACUAUUGG .....(((((((((((.(((.((....-.))))))))))).(((((........)))))............(((((......)))))...(((((((((....)))..))))))))))) ( -37.00, z-score = -3.06, R) >consensus AUUAACAAAUAUACCG_CUGAGUCUCUAGGCCAGCGGUAUAGAAGGACAGUCU_CCUUCCAACCUAAUCAACCUAUAUCAACAUAGAAGCAGGACGGCCAACCGGCU_C_CACU_____ ...........(((((.(((.(((....)))))))))))..(((((........)))))............................................................ (-13.58 = -12.37 + -1.21)

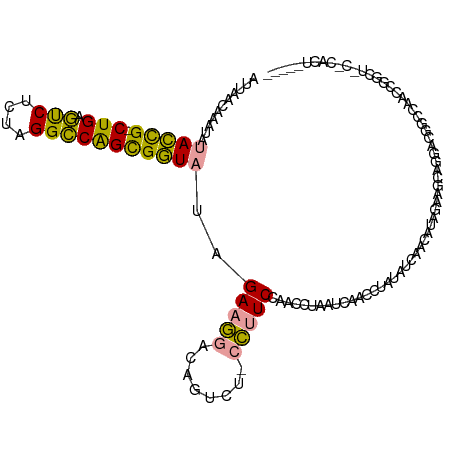
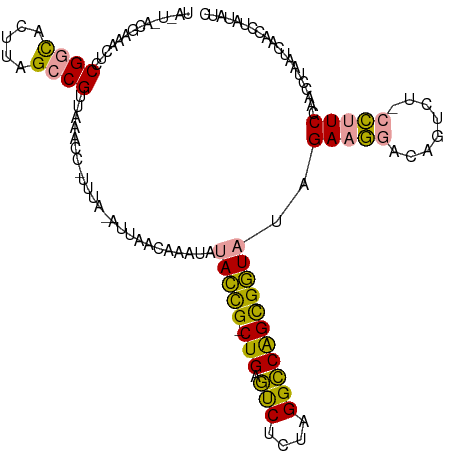
| Location | 21,664,671 – 21,664,787 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 62.25 |
| Shannon entropy | 0.53446 |
| G+C content | 0.40173 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -16.40 |
| Energy contribution | -15.30 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21664671 116 - 22422827 UA-UAACGAAACUCCGGCACUUAGCCGUUAAACCUUUUU-AUUACAAAAUAUACCGGCUGAGUCUCUAGGCCAGCGGUAUUGAAGGACAGUCU-CCUUCCAACGUAAUCAUCCUAUUUC ..-....((((...((((.....))))............-(((((.....((((((.(((.(((....)))))))))))).((((((.....)-)))))....))))).......)))) ( -28.80, z-score = -2.80, R) >droWil1.scaffold_181075 769703 111 + 1141854 ------CAUAACUCCGACACUUGGCCGUAACACAAAUUAUAAUUUCUAAUAUAUUG-CUGAUCCACAAGGACGGUGAUUCAGAUACUUAAUCGUAUCUCC-UCCUAAUCAACAUAAAUC ------.........((.....((............................((..-(((.(((....))))))..))..((((((......))))))))-......)).......... ( -13.80, z-score = -0.16, R) >droVir3.scaffold_13050 3111385 116 + 3426691 UAUUCAUGAAACUUCGGUACUUAGCCGUUAAAUC-UUUA-AUUAACCAAUAUACCGACUGAGUCUAU-GGCCAGCGGUAUCGAAGGACAGUUUCCCUUCCACCCUAAUCUCCCUAUCUC .......((((((.((((.....)))).......-....-.....((...((((((.(((.((....-.)))))))))))....))..))))))......................... ( -22.10, z-score = -1.12, R) >consensus UA_U_ACGAAACUCCGGCACUUAGCCGUUAAACC_UUUA_AUUAACAAAUAUACCG_CUGAGUCUCUAGGCCAGCGGUAUAGAAGGACAGUCU_CCUUCCAACCUAAUCAACCUAUAUC ..............((((.....))))........................(((((.(((.(((....)))))))))))..(((((........))))).................... (-16.40 = -15.30 + -1.10)

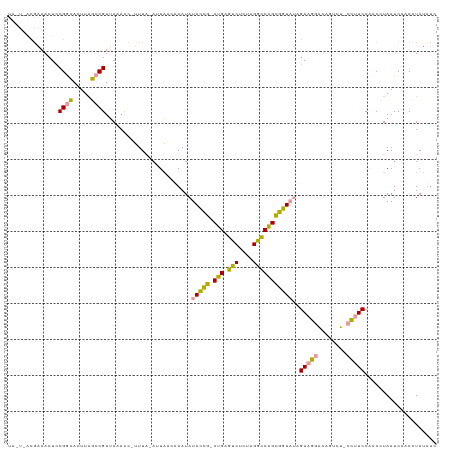
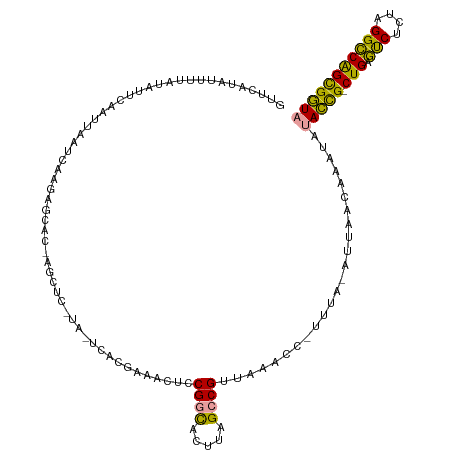
| Location | 21,664,710 – 21,664,827 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 60.85 |
| Shannon entropy | 0.54852 |
| G+C content | 0.36219 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21664710 117 - 22422827 GUUCUGAUUUUAUAUUCAAUUAAUAAAGAGCACAAGCUCAUA-UAACGAAACUCCGGCACUUAGCCGUUAAACCUUUUU-AUUACAAAAUAUACCGGCUGAGUCUCUAGGCCAGCGGUA ....(((........)))..(((((((((((....))))...-...........((((.....)))).........)))-)))).......(((((.(((.(((....))))))))))) ( -26.90, z-score = -2.47, R) >droWil1.scaffold_181075 769742 103 + 1141854 ----GUUUUGUAUAACCAAUUAAACAAUAAAG-----------ACGCAUAACUCCGACACUUGGCCGUAACACAAAUUAUAAUUUCUAAUAUAUUG-CUGAUCCACAAGGACGGUGAUU ----(((((....................)))-----------))........((((...))))............................((..-(((.(((....))))))..)). ( -9.45, z-score = 1.57, R) >droVir3.scaffold_13050 3111425 116 + 3426691 GUUCAUAUUUGAUAUUCAAUAAAUCAAGAGCACCAGCUCCUAUUCAUGAAACUUCGGUACUUAGCCGUUAAAUC-UUUA-AUUAACCAAUAUACCGACUGAGUCUAU-GGCCAGCGGUA .(((((..(((((.........)))))((((....))))......)))))....((((.....)))).......-....-...........(((((.(((.((....-.)))))))))) ( -23.50, z-score = -1.84, R) >consensus GUUCAUAUUUUAUAUUCAAUUAAUCAAGAGCAC_AGCUC_UA_UCACGAAACUCCGGCACUUAGCCGUUAAACC_UUUA_AUUAACAAAUAUACCG_CUGAGUCUCUAGGCCAGCGGUA ...........................((((....))))...............((((.....))))........................(((((.(((.(((....))))))))))) (-13.65 = -13.67 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:11 2011