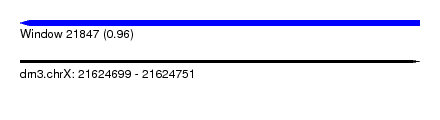
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,624,699 – 21,624,751 |
| Length | 52 |
| Max. P | 0.956435 |

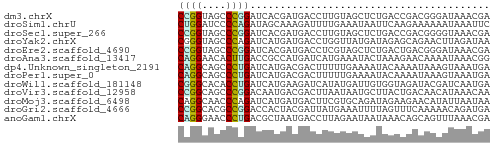
| Location | 21,624,699 – 21,624,751 |
|---|---|
| Length | 52 |
| Sequences | 13 |
| Columns | 52 |
| Reading direction | reverse |
| Mean pairwise identity | 58.60 |
| Shannon entropy | 0.94372 |
| G+C content | 0.43343 |
| Mean single sequence MFE | -8.35 |
| Consensus MFE | -4.24 |
| Energy contribution | -3.88 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
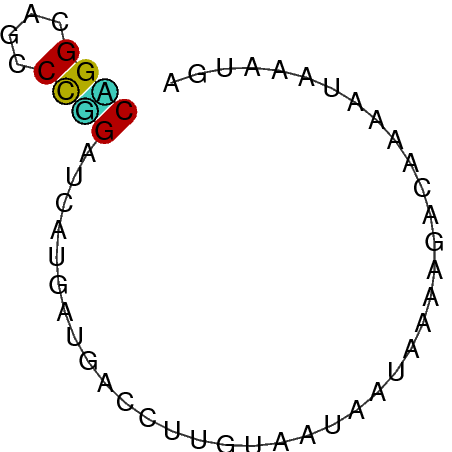
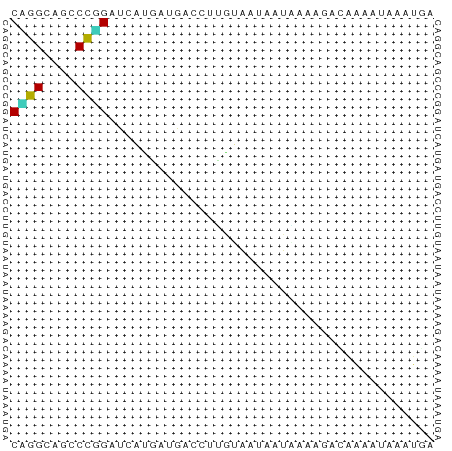
>dm3.chrX 21624699 52 - 22422827 CCGGUAGCCCGGAUCACGAUGACCUUGUAGCUCUGACCGACGGGAUAAACGA ((.((....((((.(((((.....)))).).))))....)).))........ ( -8.80, z-score = 1.35, R) >droSim1.chrU 6983292 52 + 15797150 CUGGAUCCCCAGAUAGCAAAGAUUUUGAAAUAAUUCAAGAAAAAAUAAAUUC ((((....))))..........(((((((....)))))))............ ( -8.00, z-score = -1.84, R) >droSec1.super_266 11051 52 + 17749 CCGGUAGCCCGGAUCACGAUGACCUUGUAGCUCUGACCGACGGGGUAAACGA ((((....))))....((...(((((((...........)))))))...)). ( -13.60, z-score = 0.24, R) >droYak2.chrX 21169754 52 - 21770863 CGGGUAGCCCAGAUCAUGAUGACCUGGUUAUGAUAGAGCAGAACUUAGAUAA .(((...)))..((((((((......)))))))).................. ( -8.80, z-score = -0.17, R) >droEre2.scaffold_4690 17992244 52 - 18748788 CCGGUAGCCCGGAUCACGAUGACCUCGUAGCUCUGACUGACGGGAUAAACGA (((.(((..((((.(((((.....)))).).)))).))).)))......... ( -12.10, z-score = 0.18, R) >droAna3.scaffold_13417 1270775 52 + 6960332 CAGGAACACUUGACCGCCAUGAUCAUGAAAUACUAAAGAACAAAAUAAACGG ((((....)))).(((.(((....))).....(....)...........))) ( -1.80, z-score = 0.61, R) >dp4.Unknown_singleton_2191 7428 52 + 19051 CAGGCAGCCCUGAUCAUGACGACUUUUUGAAAAUACAAAAUAAAGUAAAUGA ((((....)))).((((....((((((((......)))...)))))..)))) ( -7.50, z-score = -1.40, R) >droPer1.super_0 11752787 52 - 11822988 CAGGCAGCCCUGAUCAUGACGACUUUUUGAAAAUACAAAAUAAAGUAAAUGA ((((....)))).((((....((((((((......)))...)))))..)))) ( -7.50, z-score = -1.40, R) >droWil1.scaffold_181148 3318799 52 - 5435427 CGGGCACACCUGAUCAUGAAGAUCAUAUGAUUGUGGUAGAUACGAUCAAUGA .((.....))(((((.....)))))..((((((((.....)))))))).... ( -13.10, z-score = -1.14, R) >droVir3.scaffold_12958 2696273 52 + 3547706 CCGGCAGCCCGGACAAUGACGACUUAAUAAUGCUUACUGACAACAUAAACAA ((((....))))........................................ ( -5.20, z-score = -0.40, R) >droMoj3.scaffold_6498 2258731 52 - 3408170 CAGGCAACCCAGAUCAUGAUGACUUCGUGCAGAUAGAAGAACAUAUUAAUAA ..(....)......(((((.....)))))....................... ( -3.60, z-score = 0.92, R) >droGri2.scaffold_4666 2460 52 + 9413 CCGGCACGCCGGACCACUACGAUUAUGAAAUUUUAGUUUCAAAAACAGAUGA ((((....)))).............((((((....))))))........... ( -10.20, z-score = -1.94, R) >anoGam1.chrX 20181516 52 - 22145176 CAGGGAACCCUGACGCUAAUGACCUUAGAAUAAUAAACAGCAGUUUAAACGA ((((....)))).((((((.....)))).....(((((....)))))..)). ( -8.30, z-score = -1.63, R) >consensus CAGGCAGCCCGGAUCAUGAUGACCUUGUAAUAAUAAAAGACAAAAUAAAUGA ((((....))))........................................ ( -4.24 = -3.88 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:07 2011