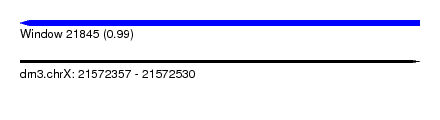
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,572,357 – 21,572,530 |
| Length | 173 |
| Max. P | 0.994723 |

| Location | 21,572,357 – 21,572,530 |
|---|---|
| Length | 173 |
| Sequences | 3 |
| Columns | 175 |
| Reading direction | reverse |
| Mean pairwise identity | 55.45 |
| Shannon entropy | 0.62099 |
| G+C content | 0.48687 |
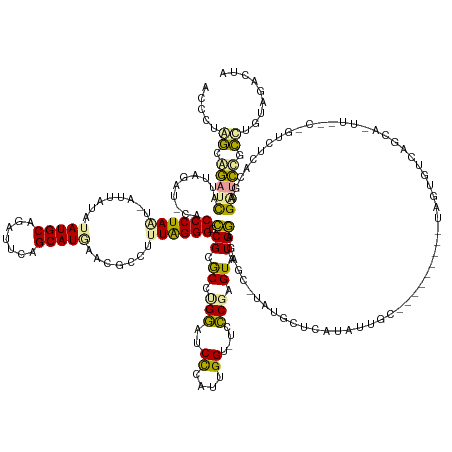
| Mean single sequence MFE | -57.53 |
| Consensus MFE | -18.86 |
| Energy contribution | -17.66 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
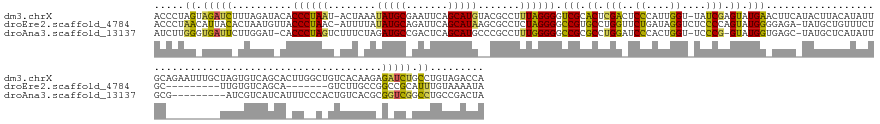
>dm3.chrX 21572357 173 - 22422827 ACCCUAGUAGAUCUUUAGAUACACCCUAAU-ACUAAAUAUGCGAAUUCAGCAUGUACGCCUUUAGGGGUCGCACUCGACUCCCAUUGGU-UAUCGAGUAUGAACUUCAUACUUACAUAUUGCAGAAUUUGCUAGUGUCAGCACUUGGCUGUCACAAGAGAUCUGCCUGUAGACCA ...(((((((((((((..............-.........((((((((.((((((..(((..(.(((((((....))))))).)..)))-....(((((((.....))))))))))...))).))))))))..(((.((((.....)))).))).))))))))))...))).... ( -56.10, z-score = -4.26, R) >droEre2.scaffold_4784 23287692 157 - 25762168 ACCCUAACAUUACACUAAUGUUACCCUAAC-AUUUUAUAUGCAGAUUCAGCAUAAGCGCCUCUAGGGGCCGUGCCUGGUUCUGAUAGGUCUCCCCAGUAUGGGGAGA-UAUGCUGUUUCUGC---------UUGUGUCAGCA-------GUCUUGCCGGCCGCAUUUGUAAAAUA ....((((((((...)))))))).......-((((((((.(((((..(((((((...((((...(((((((....)))))))...))))((((((.....)))))).-)))))))..)))))---------.((((((.((.-------.....)).)).))))..)))))))). ( -55.90, z-score = -3.25, R) >droAna3.scaffold_13137 422782 162 - 1373759 AUCUUGGGUGAUUCUUGGAU-CACCCUAGUCUUUCUAGAUGCCGACUCAGCAUGCCCGCCUUUGGGGGCCGCGCCUGGAUCCCACUGGU-UCCCG-GUAUGGUGAGC-UAUGCUCAUAUUGCG---------AUCGUCAUCAUUUCCCACUGUCACGCGGUCGGCCUGCCGACUA ..((.((((((((....)))-))))).))........((((.(((((((.((((((.(((..((((..(((....)))..))))..)))-....)-))))).)))).-..(((.......)))---------.))).)))).................((((((....)))))). ( -60.60, z-score = -1.58, R) >consensus ACCCUAGCAGAUCAUUAGAU_CACCCUAAU_AUUAUAUAUGCAGAUUCAGCAUGAACGCCUUUAGGGGCCGCGCCUGGAUCCCAUUGGU_UCCCGAGUAUGGAGAGC_UAUGCUCAUAUUGC_________UAGUGUCAGCA_UU__C_GUCUCACGAGAUCCGCCUGUAGACUA .....((.(((((..........((((((........(((((.......))))).......)))))).(((.((.(((..((....))....))).)).)))........................................................))))).))......... (-18.86 = -17.66 + -1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:06 2011