
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,508,804 – 21,508,885 |
| Length | 81 |
| Max. P | 0.999606 |

| Location | 21,508,804 – 21,508,885 |
|---|---|
| Length | 81 |
| Sequences | 3 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 54.29 |
| Shannon entropy | 0.66256 |
| G+C content | 0.43240 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -13.54 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
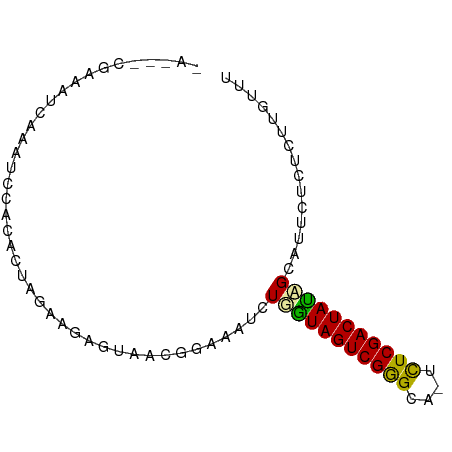
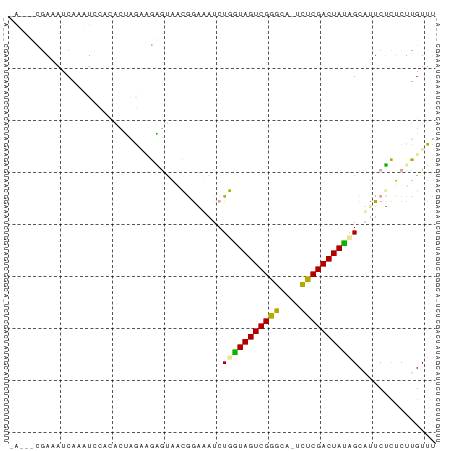
>dm3.chrX 21508804 81 + 22422827 AAUUUCGAGAUCGCAUUCACACUAGCUGAGUAACGGAUAUCUGAUAGUCGGGGAA-CUCGACUAUCGCAUUCUCUCUUGUUU ......((((..(((((((.......)))))...........((((((((((...-))))))))))))..))))........ ( -22.70, z-score = -2.08, R) >droEre2.scaffold_4929 5446611 79 - 26641161 -AGAGCGAAAUCAACUCAAAAAGAAGAGCACAACAAUCCUCUGGUAGUCGGGU--UUUCGACUAUAGCAUUCGCUCUUGUUU -((((((((.....(((........)))...............((((((((..--..))))))))....))))))))..... ( -20.90, z-score = -2.22, R) >droMoj3.scaffold_6482 1073263 76 - 2735782 ------AAUAUCAGUUGCUCGCUUGACAGGUUCAGGGAAUCUUGUAGUCGAGCAGUCUCGACUACAGCUUUCUUACUUGUUC ------..................(((((((...(((((..(((((((((((....)))))))))))..)))))))))))). ( -25.30, z-score = -3.10, R) >consensus _A___CGAAAUCAAAUCCACACUAGAAGAGUAACGGAAAUCUGGUAGUCGGGCA_UCUCGACUAUAGCAUUCUCUCUUGUUU .........................................(((((((((((....)))))))))))............... (-13.54 = -12.77 + -0.77)

| Location | 21,508,804 – 21,508,885 |
|---|---|
| Length | 81 |
| Sequences | 3 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 54.29 |
| Shannon entropy | 0.66256 |
| G+C content | 0.43240 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -9.51 |
| Energy contribution | -9.73 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
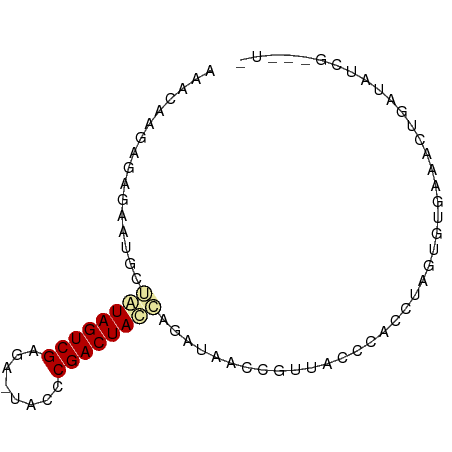
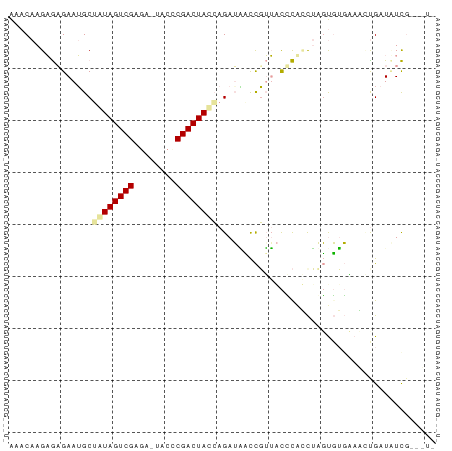
>dm3.chrX 21508804 81 - 22422827 AAACAAGAGAGAAUGCGAUAGUCGAG-UUCCCCGACUAUCAGAUAUCCGUUACUCAGCUAGUGUGAAUGCGAUCUCGAAAUU ........((((.(((((((((((..-.....)))))))).........((((.(.....).))))..))).))))...... ( -21.70, z-score = -2.48, R) >droEre2.scaffold_4929 5446611 79 + 26641161 AAACAAGAGCGAAUGCUAUAGUCGAAA--ACCCGACUACCAGAGGAUUGUUGUGCUCUUCUUUUUGAGUUGAUUUCGCUCU- .....((((((((.(((.((((((...--...))))))..((((.((....)).))))........)))....))))))))- ( -23.70, z-score = -2.95, R) >droMoj3.scaffold_6482 1073263 76 + 2735782 GAACAAGUAAGAAAGCUGUAGUCGAGACUGCUCGACUACAAGAUUCCCUGAACCUGUCAAGCGAGCAACUGAUAUU------ ..............((((((((((((....))))))))))....(((.(((.....))).).))))..........------ ( -17.90, z-score = -1.70, R) >consensus AAACAAGAGAGAAUGCUAUAGUCGAGA_UACCCGACUACCAGAUAACCGUUACCCACCUAGUGUGAAACUGAUAUCG___U_ ................((((((((........)))))))).......................................... ( -9.51 = -9.73 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:07:04 2011