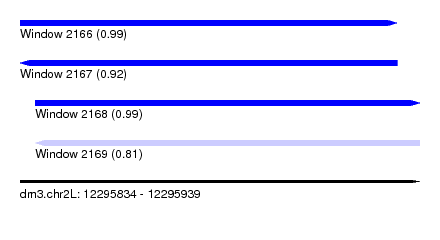
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,295,834 – 12,295,939 |
| Length | 105 |
| Max. P | 0.992067 |

| Location | 12,295,834 – 12,295,933 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.40463 |
| G+C content | 0.45610 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
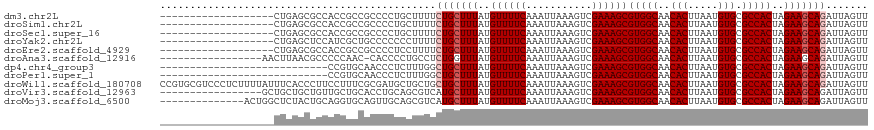
>dm3.chr2L 12295834 99 + 23011544 -------------------CUGAGCGCCACCGCCGCCCCUGCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -------------------..(.(((....))))......(((..((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))). ( -30.70, z-score = -2.79, R) >droSim1.chr2L 12096372 99 + 22036055 -------------------CUGAGCGCCACCGCCGCCCCUGCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -------------------..(.(((....))))......(((..((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))). ( -30.70, z-score = -2.79, R) >droSec1.super_16 485570 99 + 1878335 -------------------CUGAGCGCCACCGCCGCCCCUGCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -------------------..(.(((....))))......(((..((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))). ( -30.70, z-score = -2.79, R) >droYak2.chr2L 8726344 99 + 22324452 -------------------CUGAGCUCCAUCGCUGCCCCCCCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -------------------...(((......))).......((..((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..)).. ( -29.30, z-score = -3.49, R) >droEre2.scaffold_4929 13518891 99 - 26641161 -------------------CUGAGCGCCACCGCCGCCCCUCCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -------------------..(.(((....)))).......((..((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..)).. ( -29.30, z-score = -2.99, R) >droAna3.scaffold_12916 3079926 100 - 16180835 -----------------AACUUAACGCCCCCAAC-CACCCCUGCCUCUGGUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -----------------.................-.....((((.((((((..((((((((...........))))))))(((..(((.....))).))))))))).))))....... ( -25.20, z-score = -2.54, R) >dp4.chr4_group3 5870552 90 - 11692001 ----------------------------CCGUGCAACCCUCUUUGGCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU ----------------------------.........((.....))(((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....... ( -24.30, z-score = -1.55, R) >droPer1.super_1 2973008 90 - 10282868 ----------------------------CCGUGCAACCCUCUUUGGCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU ----------------------------.........((.....))(((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....... ( -24.30, z-score = -1.55, R) >droWil1.scaffold_180708 9991168 118 + 12563649 CCGUGCGUCCCUCUUUUAUUUCACCCUUCCUUUCGCGAUGCUGCUGCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU ....(((((.(.......................).))))).(((((((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))..)))). ( -28.00, z-score = -1.07, R) >droVir3.scaffold_12963 17375707 101 + 20206255 -----------------GCUGCUGCUGUUGCUGCACCUGCAGCGUCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU -----------------.(((((((((.((((((....)))))).)).)).....((((((...........))))))(((((..(((.....))).)))))....)))))....... ( -33.80, z-score = -1.62, R) >droMoj3.scaffold_6500 22425648 104 + 32352404 --------------ACUGGCUCUACUGCAGGUGCAGUUGCAGCGUCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU --------------((((((((.(((((....))))).).)))(((.((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))))))). ( -34.00, z-score = -1.71, R) >consensus ___________________CUGAGCGCCACCGCCGCCCCUCCUUUUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUU ..............................................(((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....... (-22.36 = -22.64 + 0.27)
| Location | 12,295,834 – 12,295,933 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.40463 |
| G+C content | 0.45610 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12295834 99 - 23011544 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGCAGGGGCGGCGGUGGCGCUCAG------------------- ......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))))).......(((((.(...).)))))..------------------- ( -31.70, z-score = -1.86, R) >droSim1.chr2L 12096372 99 - 22036055 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGCAGGGGCGGCGGUGGCGCUCAG------------------- ......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))))).......(((((.(...).)))))..------------------- ( -31.70, z-score = -1.86, R) >droSec1.super_16 485570 99 - 1878335 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGCAGGGGCGGCGGUGGCGCUCAG------------------- ......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))))).......(((((.(...).)))))..------------------- ( -31.70, z-score = -1.86, R) >droYak2.chr2L 8726344 99 - 22324452 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGGGGGGGCAGCGAUGGAGCUCAG------------------- ..(((..((((((((.(((((((((.....))).))))))..((((..(((((...........)))))..))))...))))))))...))).......------------------- ( -28.60, z-score = -1.57, R) >droEre2.scaffold_4929 13518891 99 + 26641161 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGGAGGGGCGGCGGUGGCGCUCAG------------------- ..((..(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))))..))...(((((.(...).)))))..------------------- ( -32.20, z-score = -2.16, R) >droAna3.scaffold_12916 3079926 100 + 16180835 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAACCAGAGGCAGGGGUG-GUUGGGGGCGUUAAGUU----------------- (((((.(((((((((.(((((((((.....))).))))))(.((((((.......)))))).).......)))))))))..))-)))..............----------------- ( -32.00, z-score = -2.25, R) >dp4.chr4_group3 5870552 90 + 11692001 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGCCAAAGAGGGUUGCACGG---------------------------- ................(((((((((.....))).))))))(.((((((.......)))))).).....((((((......))))))....---------------------------- ( -25.10, z-score = -1.73, R) >droPer1.super_1 2973008 90 + 10282868 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGCCAAAGAGGGUUGCACGG---------------------------- ................(((((((((.....))).))))))(.((((((.......)))))).).....((((((......))))))....---------------------------- ( -25.10, z-score = -1.73, R) >droWil1.scaffold_180708 9991168 118 - 12563649 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGCAGCAGCAUCGCGAAAGGAAGGGUGAAAUAAAAGAGGGACGCACGG ........(((.(((.(((((((((.....))).)))))).((((((.(((((...........)))))..((....))....))))))...................))).)))... ( -26.10, z-score = 0.62, R) >droVir3.scaffold_12963 17375707 101 - 20206255 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGACGCUGCAGGUGCAGCAACAGCAGCAGC----------------- .......(((((....(((((((((.....))).))))))(.((((((.......)))))).).....((.((..(((((....)))))..))))))))).----------------- ( -32.70, z-score = -1.81, R) >droMoj3.scaffold_6500 22425648 104 - 32352404 AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGACGCUGCAACUGCACCUGCAGUAGAGCCAGU-------------- ......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))).)).(((.(.(((((....))))).))))....-------------- ( -30.20, z-score = -1.69, R) >consensus AACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAAAAGGAGGGGCGGAGGUGGCGCUCAG___________________ ......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))))............................................. (-21.48 = -21.93 + 0.45)

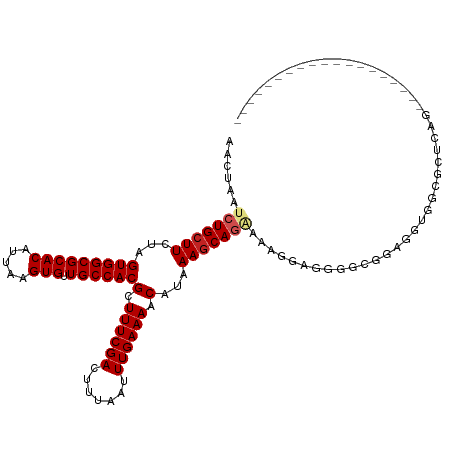
| Location | 12,295,838 – 12,295,939 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 124 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.32857 |
| G+C content | 0.44439 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
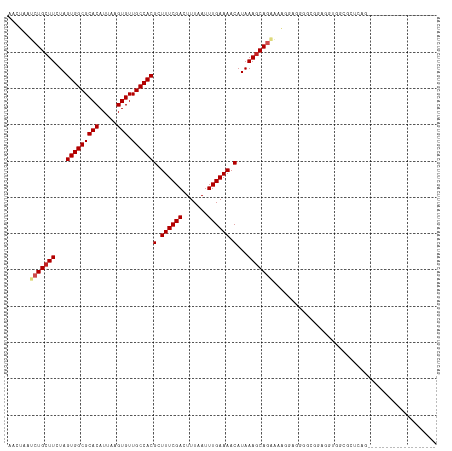
>dm3.chr2L 12295838 101 + 23011544 ---------------------GCGCCACCGCCGCCCCUGCUUUUC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------(((....))).((....(((..((--((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))).)).... ( -32.60, z-score = -3.30, R) >droSim1.chr2L 12096376 101 + 22036055 ---------------------GCGCCACCGCCGCCCCUGCUUUUC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------(((....))).((....(((..((--((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))).)).... ( -32.60, z-score = -3.30, R) >droSec1.super_16 485574 101 + 1878335 ---------------------GCGCCACCGCCGCCCCUGCUUUUC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------(((....))).((....(((..((--((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))).)).... ( -32.60, z-score = -3.30, R) >droYak2.chr2L 8726348 101 + 22324452 ---------------------GCUCCAUCGCUGCCCCCCCUUUUC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------((......))(((.....((..((--((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))..)))... ( -28.90, z-score = -3.35, R) >droEre2.scaffold_4929 13518895 101 - 26641161 ---------------------GCGCCACCGCCGCCCCUCCUUUUC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------(((....))).((.....((..((--((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))..)).... ( -29.70, z-score = -3.07, R) >droAna3.scaffold_12916 3079932 100 - 16180835 ---------------------ACGCCCCCAAC-CACCCCUGCCUC--UGGUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------.......((((-.....((((.((--((((..((((((((...........))))))))(((..(((.....))).))))))))).))))....))))..... ( -26.80, z-score = -2.59, R) >dp4.chr4_group3 5870555 93 - 11692001 -----------------------------UGCAACCCUCUUUGGC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA -----------------------------((((((((.....))(--((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....))))))... ( -31.50, z-score = -3.52, R) >droPer1.super_1 2973011 93 - 10282868 -----------------------------UGCAACCCUCUUUGGC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA -----------------------------((((((((.....))(--((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....))))))... ( -31.50, z-score = -3.52, R) >droWil1.scaffold_180708 9991172 120 + 12563649 GCGUCCCUCUUUUAUUUCACCCUUCCUUUCGCGAUGCUGCU--GC--UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA (((..........................)))(((((.(((--((--((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))..)))).))))). ( -33.67, z-score = -2.78, R) >droVir3.scaffold_12963 17375711 103 + 20206255 ---------------------CUGCUGUUGCUGCACCUGCAGCGUCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ---------------------.(((....(((((....)))))(((.((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))).....)))... ( -33.70, z-score = -1.86, R) >droMoj3.scaffold_6500 22425652 106 + 32352404 ------------------GCUCUACUGCAGGUGCAGUUGCAGCGUCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ------------------.....(((((....)))))(((((((((.((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))))..))))))... ( -36.10, z-score = -2.30, R) >consensus _____________________GCGCCACCGCCGCCCCUGCUUUUC__UGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA ...............................................((((((..((((((...........))))))(((((..(((.....))).)))))..)))))).............. (-20.26 = -20.35 + 0.09)
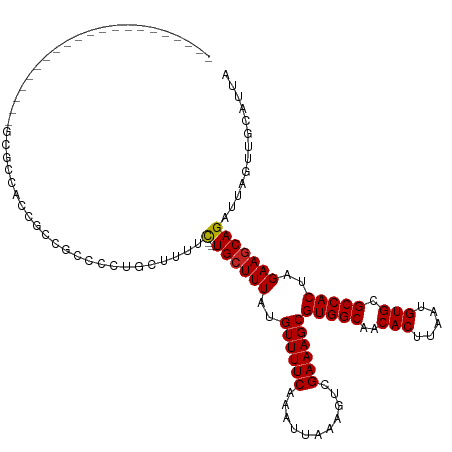
| Location | 12,295,838 – 12,295,939 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.32857 |
| G+C content | 0.44439 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.31 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12295838 101 - 23011544 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GAAAAGCAGGGGCGGCGGUGGCGC--------------------- ....((.(((...((((((((.(((((((((.....))).))))))((((((..(((((...........)))))..--).))))))))))))).))).))..--------------------- ( -34.30, z-score = -2.58, R) >droSim1.chr2L 12096376 101 - 22036055 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GAAAAGCAGGGGCGGCGGUGGCGC--------------------- ....((.(((...((((((((.(((((((((.....))).))))))((((((..(((((...........)))))..--).))))))))))))).))).))..--------------------- ( -34.30, z-score = -2.58, R) >droSec1.super_16 485574 101 - 1878335 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GAAAAGCAGGGGCGGCGGUGGCGC--------------------- ....((.(((...((((((((.(((((((((.....))).))))))((((((..(((((...........)))))..--).))))))))))))).))).))..--------------------- ( -34.30, z-score = -2.58, R) >droYak2.chr2L 8726348 101 - 22324452 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GAAAAGGGGGGGCAGCGAUGGAGC--------------------- ...(((..((..(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))--)).....))..)))((......))--------------------- ( -29.80, z-score = -2.02, R) >droEre2.scaffold_4929 13518895 101 + 26641161 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GAAAAGGAGGGGCGGCGGUGGCGC--------------------- ....((.(((...((((((((.(((((((((.....))).))))))..((((..(((((...........)))))..--))))...)))))))).))).))..--------------------- ( -31.10, z-score = -1.79, R) >droAna3.scaffold_12916 3079932 100 + 16180835 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAACCA--GAGGCAGGGGUG-GUUGGGGGCGU--------------------- ...(.((((((.(((((((((.(((((((((.....))).))))))(.((((((.......)))))).).......)--))))))))..))-)))).).....--------------------- ( -35.50, z-score = -3.32, R) >dp4.chr4_group3 5870555 93 + 11692001 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GCCAAAGAGGGUUGCA----------------------------- ...((((((....((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))--)((.....))))))))----------------------------- ( -29.50, z-score = -2.88, R) >droPer1.super_1 2973011 93 + 10282868 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GCCAAAGAGGGUUGCA----------------------------- ...((((((....((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))--)((.....))))))))----------------------------- ( -29.50, z-score = -2.88, R) >droWil1.scaffold_180708 9991172 120 - 12563649 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA--GC--AGCAGCAUCGCGAAAGGAAGGGUGAAAUAAAAGAGGGACGC ...(((..((...((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))--).--))..)))((.(....)))....................... ( -26.80, z-score = 0.43, R) >droVir3.scaffold_12963 17375711 103 - 20206255 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGACGCUGCAGGUGCAGCAACAGCAG--------------------- ...(((......(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))).)).(((((....)))))....))).--------------------- ( -32.70, z-score = -2.22, R) >droMoj3.scaffold_6500 22425652 106 - 32352404 UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGACGCUGCAACUGCACCUGCAGUAGAGC------------------ ...((((.(...(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))).)).).))))(((((....))))).....------------------ ( -30.00, z-score = -1.62, R) >consensus UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCA__GAAAAGCAGGGGCGGCGGUGGCGC_____________________ ..............(((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))............................................... (-18.22 = -18.31 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:48 2011