| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,484,720 – 21,484,834 |
| Length | 114 |
| Max. P | 0.899020 |

| Location | 21,484,720 – 21,484,834 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.64394 |
| G+C content | 0.58232 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.899020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
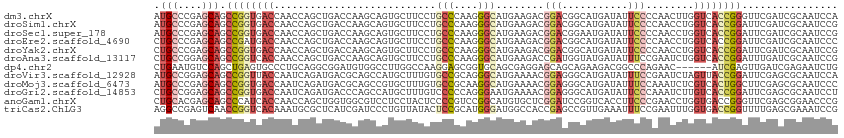
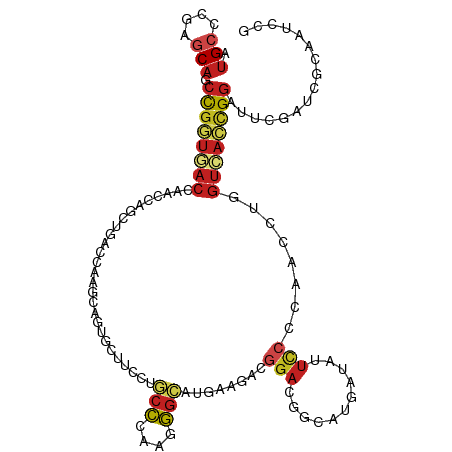
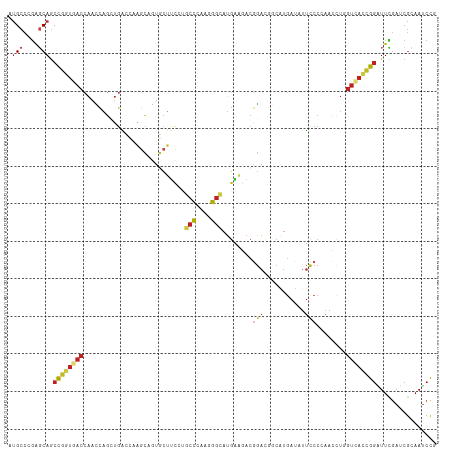
>dm3.chrX 21484720 114 - 22422827 AUGCCCGAGCAGCCGGUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACUUGGUCACCGGGUUCGAUCGCAAUCCA .(((.(((((..(((((((((((((.(((.....))).((.((((.((((....)))).))))))))..((...........))...))))))))))))))))...)))..... ( -46.20, z-score = -3.05, R) >droSim1.chrX 16554866 114 - 17042790 AUGCCCGAGCAGCCGGUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG .(((.((((...((((((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).)))))))))))).))))...)))..... ( -44.10, z-score = -3.15, R) >droSec1.super_178 10213 114 - 58033 AUGCCCGAGCAGCCGGUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGAAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG .(((.((((...((((((((((.((.(((.....))).((.((((.((((....)))).))))))))..(((((...)))))......)))))))))).))))...)))..... ( -46.80, z-score = -4.23, R) >droEre2.scaffold_4690 17955722 114 - 18748788 CUGCCCGAGCAGCCGAUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCC .(((.((((...(((.((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).)))))))).))).))))...)))..... ( -37.30, z-score = -1.86, R) >droYak2.chrX 20615021 114 - 21770863 CUGCCCGAGCAGCCGGUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG .(((.((((...((((((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).)))))))))))).))))...)))..... ( -43.90, z-score = -3.07, R) >droAna3.scaffold_13117 2961828 114 - 5790199 CUGCCGGAGCAGCCGGUCACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACCGAUGGUAUGAUAUUUCCGAAUCUGGUCACCGGAUUUGAUCGCAAUCCG .(((.((((((((.(((.....))).))))((((..(.((.((((.((((....)))).)))))).).)))).......))))((((((((...))))))))....)))..... ( -38.80, z-score = -1.62, R) >dp4.chr2 2858825 108 + 30794189 CUGAAUGUCCAGCUGAGUGCCCUGCAGGCGGAUGUGGCCUUGGCCAAGGAGCGGUGCAGCGAGGAGCAGCAGAAGACGGCCCAGAAC------AUCGAGUUGAUCGAGAAUCUG (((....(((.(((((.(((((((((......)))(((....))).))).))).).))))..)))....))).........((((..------.((((.....))))...)))) ( -34.00, z-score = 0.74, R) >droVir3.scaffold_12928 1310165 114 + 7717345 AUGCCGGAGCAGCCGGUUACCAAUCAGAUGACGCAGCCAUGCUUUGUGCCGCAGGGCAUGAAAACGGAGGGCAUGAUAUUUCCGAAUCUAGUUACCGGAUUCGAGCGCAAUCCA ..((((((....(((((....(((.((((........(((((((((.....)))))))))....((((((........)))))).)))).)))))))).)))).))........ ( -34.80, z-score = -0.05, R) >droMoj3.scaffold_6473 13958806 114 - 16943266 AUGCCCGAGCAGCCGGUGACCAAUCAGAUGACGCAGCCGUGCUUUGUGCCGCAAGGCAUGAAAACGGAGGGCAUGAUAUUUCCAAAUCUCGUCACUGGCUUCGAGCGCAAUCCC .(((((((..((((((((((............((..((((..((..((((....))))..)).))))...))..(((........)))..))))))))))))).).)))..... ( -46.30, z-score = -3.10, R) >droGri2.scaffold_14853 1808062 114 + 10151454 CUGCCGGAGCAGCCGGUGACCAAUCAGAUGACCCAGCCAUGCUUUGUCCCCCAGGGAAUGAAAACGGAGGGCAUGAUAUUCCCAAAUCUUGUCACCGGAUUCGAGCGCAAUCCU ..((((((....((((((((.......(((.(((..((........((((...))))........)).))))))(((........)))..)))))))).)))).))........ ( -33.59, z-score = -0.02, R) >anoGam1.chrX 18058618 114 + 22145176 CUGCACGAGCAGCCCAUCACCAACCAGCUGGUGGCGUCCUCCUACUCCCCGUCCGGCAUGUGCUCGGAUCCGGUCACCUUCCCGAACCUGGUGACCGGGUUCGAGCGGAACCCG ((((....))))...........((.((((((((.(..........).))).)))))....(((((((((((((((((...........)))))))))))))))))))...... ( -50.50, z-score = -3.24, R) >triCas2.ChLG3 1011543 114 - 32080666 AGGCCGAGUCAACCGGUCACAAAUGCGCUCAUCGAUCCCUGUUAUACUCCGCAUGGGAUGGCCACCGAGCCGUUGAAAUUUCCGAAUUUGGUGACCGGUUUUGAGCGAAAUCCG ..(((((...((((((((((.((((.((((..(.((((((((........))).))))).).....)))))))).(((((....))))).))))))))))))).))........ ( -36.90, z-score = -1.20, R) >consensus AUGCCCGAGCAGCCGGUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG .(((....))).((((((((...........................(((....)))........(((...........)))........))))))))................ (-13.80 = -14.53 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:59 2011