| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,478,628 – 21,478,727 |
| Length | 99 |
| Max. P | 0.671422 |

| Location | 21,478,628 – 21,478,727 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.48084 |
| G+C content | 0.55464 |
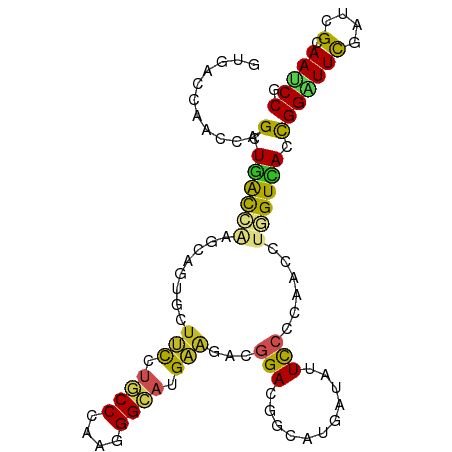
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -14.30 |
| Energy contribution | -13.95 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
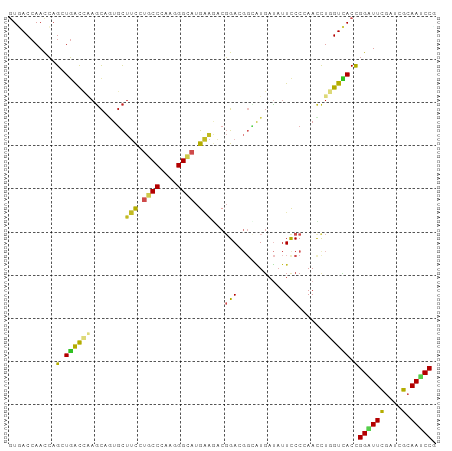
>dm3.chrX 21478628 99 - 22422827 GUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACUUGGUCACCGGGUUCGAUCGCAAUCCA (((((.((((.(.((((((((..((.((((.((((....)))).))))))(((...........)))....)))))))).).)))).).))))...... ( -35.10, z-score = -2.01, R) >droSim1.chrX 16554866 99 - 17042790 GUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG (((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).)))))))))(((((((....).)))))) ( -34.80, z-score = -2.62, R) >droSec1.super_178 10213 99 - 58033 GUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGAAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG (((((((.((.(((.....))).((.((((.((((....)))).))))))))..(((((...)))))......)))))))(((((((....).)))))) ( -37.50, z-score = -3.70, R) >droEre2.scaffold_4690 17955722 99 - 18748788 AUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCC .((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).))))))))..((((((....).))))). ( -30.50, z-score = -1.51, R) >droYak2.chrX 20615021 99 - 21770863 GUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG (((((((.((.(((.....))).((.((((.((((....)))).))))))))..((..((........)).)))))))))(((((((....).)))))) ( -34.80, z-score = -2.62, R) >droAna3.scaffold_13117 2961828 99 - 5790199 GUCACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACCGAUGGUAUGAUAUUUCCGAAUCUGGUCACCGGAUUUGAUCGCAAUCCG ((((...(((((((.....))).(..((((.((((....)))).))))..)..)))).)))).....(((((((((...)))))))))........... ( -30.80, z-score = -1.87, R) >droVir3.scaffold_12928 1310165 99 + 7717345 GUUACCAAUCAGAUGACGCAGCCAUGCUUUGUGCCGCAGGGCAUGAAAACGGAGGGCAUGAUAUUUCCGAAUCUAGUUACCGGAUUCGAGCGCAAUCCA ...........(((..(((...(((((((((.....))))))))).....(((((........)))))((((((.......))))))..)))..))).. ( -27.80, z-score = -0.44, R) >droMoj3.scaffold_6473 13958806 99 - 16943266 GUGACCAAUCAGAUGACGCAGCCGUGCUUUGUGCCGCAAGGCAUGAAAACGGAGGGCAUGAUAUUUCCAAAUCUCGUCACUGGCUUCGAGCGCAAUCCC (((..(((((((.((((((..((((..((..((((....))))..)).))))...))..(((........)))..)))))))).)).)..)))...... ( -33.30, z-score = -1.55, R) >droGri2.scaffold_14853 1808062 99 + 10151454 GUGACCAAUCAGAUGACCCAGCCAUGCUUUGUCCCCCAGGGAAUGAAAACGGAGGGCAUGAUAUUCCCAAAUCUUGUCACCGGAUUCGAGCGCAAUCCU (((((.......(((.(((..((........((((...))))........)).))))))(((........)))..))))).((((((....).))))). ( -21.59, z-score = 1.14, R) >anoGam1.chrX 18058618 99 + 22145176 AUCACCAACCAGCUGGUGGCGUCCUCCUACUCCCCGUCCGGCAUGUGCUCGGAUCCGGUCACCUUCCCGAACCUGGUGACCGGGUUCGAGCGGAACCCG ........((.((((((((.(..........).))).)))))....(((((((((((((((((...........)))))))))))))))))))...... ( -46.60, z-score = -3.90, R) >triCas2.ChLG3 1011543 99 - 32080666 GUCACAAAUGCGCUCAUCGAUCCCUGUUAUACUCCGCAUGGGAUGGCCACCGAGCCGUUGAAAUUUCCGAAUUUGGUGACCGGUUUUGAGCGAAAUCCG ..........((((((...((((((((........))).)))))((.(((((((...(((.......))).))))))).)).....))))))....... ( -28.30, z-score = -0.87, R) >consensus GUGACCAACCAGCUGACCAAGCAGUGCUUCCUGCCCAAGGGCAUGAAGACGGACGGCAUGAUAUUCCCCAACCUGGUCACCGGAUUCGAUCGCAAUCCG ...........(.((((((........(((.((((....)))).)))...(((...........)))......)))))).)((((((....).))))). (-14.30 = -13.95 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:58 2011