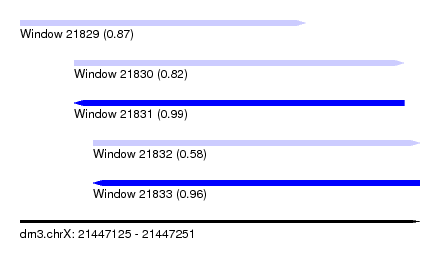
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,447,125 – 21,447,251 |
| Length | 126 |
| Max. P | 0.993361 |

| Location | 21,447,125 – 21,447,215 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.61 |
| Shannon entropy | 0.35595 |
| G+C content | 0.51835 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -17.97 |
| Energy contribution | -20.29 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21447125 90 + 22422827 UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACUGAACUGAGUUCAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCC------- .....(((((((((((((...)))))(((((.....(((((((((((((((....)))))))))))))))))))).....))))))))..------- ( -44.90, z-score = -5.82, R) >droAna3.scaffold_13117 2798910 69 - 5790199 UUACUGUCGAUGUGCGGUUAAAUUGCGGUUCGUUCUUGGCC--------------CAAUUCGAU-------GGCCAAAAUGUCCUCUUCC------- ...(((((((..((.((((((...(((...)))..))))))--------------))..)))))-------)).................------- ( -15.10, z-score = -0.62, R) >droEre2.scaffold_4690 17928266 80 + 18748788 UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUGGCCCAGGACCAAAAAGCGUUGGCUC------- .....(((((((((((((...)))))(((((......(((((.((----------......)))))))..))))).....))))))))..------- ( -25.70, z-score = -0.87, R) >droYak2.chrX 20585056 87 + 21770863 UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCACUUGGCCAGGACCGAAAAGCGUUGGCCCUUGGCCC .....(((((...(.(((..(...((..((((.((((((((((..----------........)))))))))).))))..)).)..))))))))).. ( -29.10, z-score = -0.61, R) >droSec1.super_8 3732434 79 + 3762037 UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACC-AAAAGCAUUGGGCC------- ......((((((((((((...)))))(((((.....(((((((((----------......))))))))))))))-....)))))))...------- ( -28.40, z-score = -1.78, R) >droSim1.chrX 16531297 80 + 17042790 UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCC------- .....(((((((((((((...)))))(((((.....(((((((((----------......)))))))))))))).....))))))))..------- ( -31.30, z-score = -2.49, R) >consensus UUGUUGUCGAUGUGCGGUUGAAUUGCGGUUCGCUUUUGGCCAACU__________CAGUUCAGUUGGCCAGGACCAAAAAGCAUUGGCCC_______ .....(((((((((((((...)))))(((((.....(((((((((................)))))))))))))).....))))))))......... (-17.97 = -20.29 + 2.32)

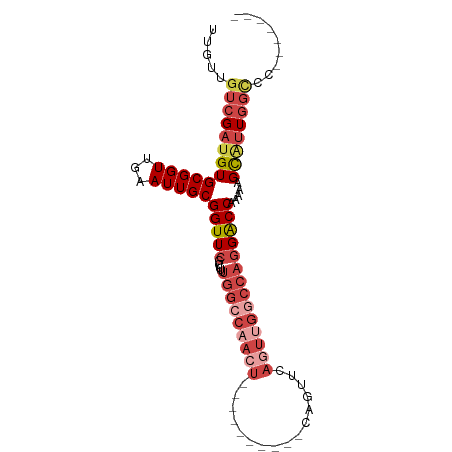
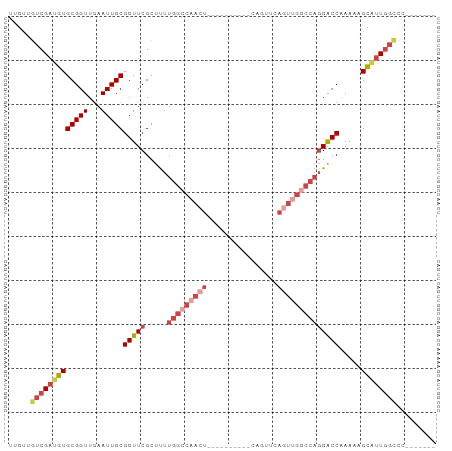
| Location | 21,447,142 – 21,447,246 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.28 |
| Shannon entropy | 0.39393 |
| G+C content | 0.55454 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -20.07 |
| Energy contribution | -19.79 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815302 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 21447142 104 + 22422827 UUGAAUUGCGGUUCGCUUUUGGCCAACUGAACUGAGUUCAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCCGU----UGGU--GG--CGGGUGG--GUGGUGCCUGUGUUUG .........(((((.....(((((((((((((((....))))))))))))))))))))...(((((((.((((((----....--.)--))))).)--...))))))....... ( -51.50, z-score = -4.90, R) >droEre2.scaffold_4690 17928283 86 + 18748788 UUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUGGCCCAGGACCAAAAAGCGUUGGCUCGU----CGGAUGGG--UGGGCUGU-GUUUG----------- .(((((((.((((.((.....)).))))----------)))))))(..(((((...(((....(((......)))----....))).--)))))..)-.....----------- ( -27.50, z-score = -0.17, R) >droYak2.chrX 20585073 104 + 21770863 UUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCACUUGGCCAGGACCGAAAAGCGUUGGCCCUUGGCCCGUGUUGGCUUGGGUGGCUGGGUGUUCUGUGUUUG .......((((..(((((..(((((.((----------((((..((..(((((((.((((......)))).)).)))))...))..)).))))))))))))))..))))..... ( -39.20, z-score = -1.22, R) >droSec1.super_8 3732451 95 + 3762037 UUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACCAAAA-GCAUUGGGCCGU----CGGU--GG--UGGCCCGUCGUUUGGCCAGUGUUUG .........(((((.....(((((((((----------......))))))))))))))..((-(((((((.(((.----((..--((--....))..))..)))))))))))). ( -39.70, z-score = -2.66, R) >droSim1.chrX 16531314 96 + 17042790 UUGAAUUGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCCGU----CGGU--GG--UGGGUGGGUGGUUGGCCAGUGUUUG .........(((((.....(((((((((----------......))))))))))))))...(((((((((((..(----((.(--.(--....).).)))..))))))))))). ( -38.00, z-score = -1.74, R) >consensus UUGAAUUGCGGUUCGCUUUUGGCCAACU__________CAGUUCAGUUGGCCAGGACCAAAAAGCAUUGGCCCGU____CGGU__GG__UGGGUGGU_GUUUGGCCAGUGUUUG ......(((..((.(.((((((((((((................)))))))))))).).))..)))...(((((...............))))).................... (-20.07 = -19.79 + -0.28)

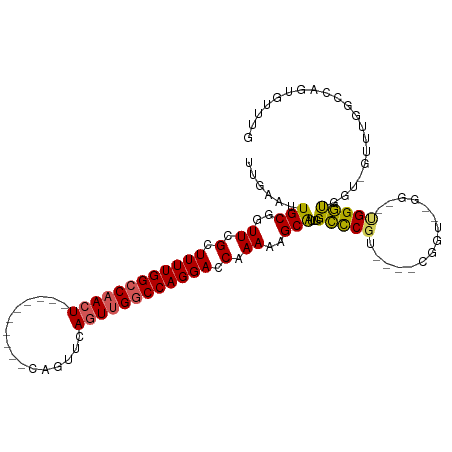
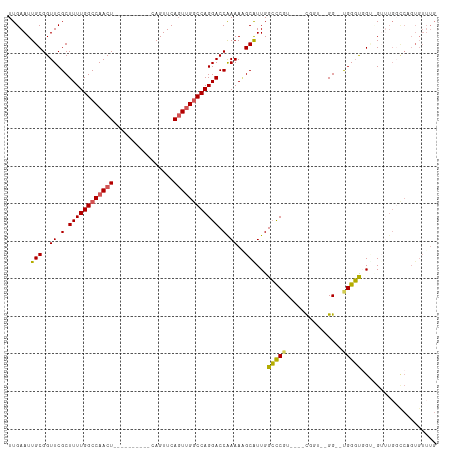
| Location | 21,447,142 – 21,447,246 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Shannon entropy | 0.39393 |
| G+C content | 0.55454 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -19.09 |
| Energy contribution | -20.29 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993361 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 21447142 104 - 22422827 CAAACACAGGCACCAC--CCACCCG--CC--ACCA----ACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUGAACUCAGUUCAGUUGGCCAAAAGCGAACCGCAAUUCAA ........(((.....--......)--))--....----..(((((((......)))))))(((((((((((((((....)))))))))))))))...(((...)))....... ( -44.40, z-score = -6.38, R) >droEre2.scaffold_4690 17928283 86 - 18748788 -----------CAAAC-ACAGCCCA--CCCAUCCG----ACGAGCCAACGCUUUUUGGUCCUGGGCCACUGAACUG----------AGUUGGCCAAAAGCGAACCGCAAUUCAA -----------.....-...(((((--.....(((----(.((((....)))).))))...)))))...((((.((----------.(((.((.....)).)))..)).)))). ( -23.70, z-score = -1.46, R) >droYak2.chrX 20585073 104 - 21770863 CAAACACAGAACACCCAGCCACCCAAGCCAACACGGGCCAAGGGCCAACGCUUUUCGGUCCUGGCCAAGUGAACUG----------AGUUGGCCAAAAGCGAACCGCAAUUCAA ........(((...............((((((...(((((.(((((..........)))))))))).((....)).----------.)))))).....(((...)))..))).. ( -31.20, z-score = -1.36, R) >droSec1.super_8 3732451 95 - 3762037 CAAACACUGGCCAAACGACGGGCCA--CC--ACCG----ACGGCCCAAUGC-UUUUGGUCCUGGCCAACUGAACUG----------AGUUGGCCAAAAGCGAACCGCAAUUCAA ........(((((((.(..(((((.--..--....----..)))))....)-.))))))).(((((((((......----------)))))))))...(((...)))....... ( -34.50, z-score = -3.03, R) >droSim1.chrX 16531314 96 - 17042790 CAAACACUGGCCAACCACCCACCCA--CC--ACCG----ACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUG----------AGUUGGCCAAAAGCGAACCGCAAUUCAA .......((((((((((..((.(((--..--((((----(.((((....)))).)))))..))).....))...))----------.))))))))...(((...)))....... ( -29.10, z-score = -2.12, R) >consensus CAAACACAGGCCAAAC_ACCACCCA__CC__ACCG____ACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUG__________AGUUGGCCAAAAGCGAACCGCAAUUCAA .........................................(((((((......)))))))(((((((((................)))))))))...(((...)))....... (-19.09 = -20.29 + 1.20)

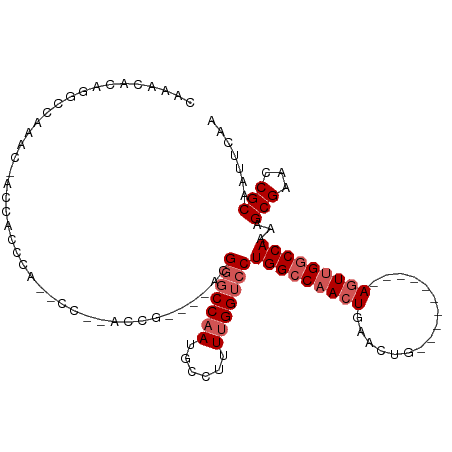
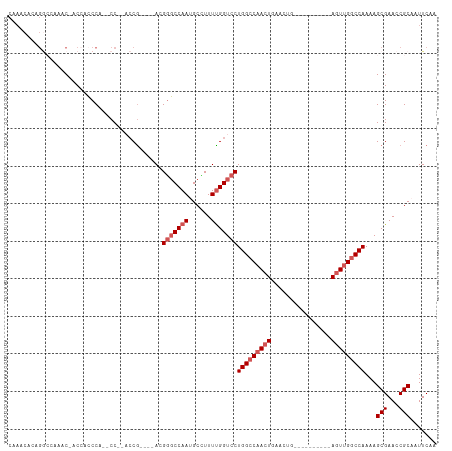
| Location | 21,447,148 – 21,447,251 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.36298 |
| G+C content | 0.59175 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -21.87 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581540 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 21447148 103 + 22422827 UGCGGUUCGCUUUUGGCCAACUGAACUGAGUUCAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCCGUUGGUGGCG--------GGUGGGUGGU--GCCUGUGUUUGCGCAC ((((...(((((((((((((((((((((....)))))))))))))))))).....(((((((.((((((.....)))--------))).)...))--)))))))....)))). ( -54.60, z-score = -4.82, R) >droEre2.scaffold_4690 17928289 85 + 18748788 UGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUGGCCCAGGACCAAAAAGCGUUGGCUCGUCGG--AUG--------GGUGGG--------CUGUGUUUGCGCAC ((((((((......(((((.((----------......)))))))..))))).....(((.((((((..(..--...--------)..)))--------)))....)))))). ( -27.90, z-score = 0.52, R) >droYak2.chrX 20585079 103 + 21770863 UGCGGUUCGCUUUUGGCCAACU----------CAGUUCACUUGGCCAGGACCGAAAAGCGUUGGCCCUUGGCCCGUGUUGGCUUGGGUGGCUGGGUGUUCUGUGUUUGCGCAC (((((....))....((.(((.----------(((..((((..((((...((((..((((..((((...))))..))))...)))).))))..))))..))).))).))))). ( -41.50, z-score = -0.82, R) >droSec1.super_8 3732457 94 + 3762037 UGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACCAAAAG-CAUUGGGCCGUCGGUGGUG--------GCCCGUCGUUUGGCCAGUGUUUGCGCAC ((((((((.....(((((((((----------......))))))))))))))..(((-((((((.(((.((..((..--------..))..))..))))))))))))..))). ( -42.60, z-score = -2.68, R) >droSim1.chrX 16531320 95 + 17042790 UGCGGUUCGCUUUUGGCCAACU----------CAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCCGUCGGUGGUG--------GGUGGGUGGUUGGCCAGUGUUUGCGCAC ((((((((.....(((((((((----------......))))))))))))))...(((((((((((..(((.(.(..--------..).).)))..)))))))))))..))). ( -40.90, z-score = -1.78, R) >consensus UGCGGUUCGCUUUUGGCCAACU__________CAGUUCAGUUGGCCAGGACCAAAAGGCAUUGGCCCGUCGGUGGUG________GGUGGGUGGUUGGCCUGUGUUUGCGCAC ((((...(((...(((((((((................)))))))))((.((((......)))).))..................................)))....)))). (-21.87 = -22.15 + 0.28)

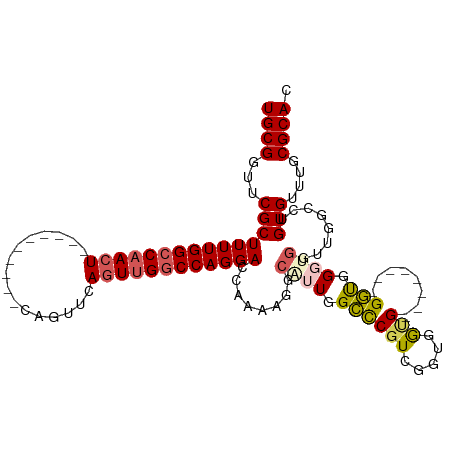
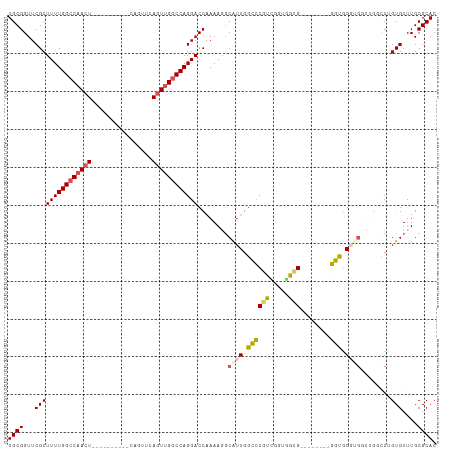
| Location | 21,447,148 – 21,447,251 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.36298 |
| G+C content | 0.59175 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -22.59 |
| Energy contribution | -23.79 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958855 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 21447148 103 - 22422827 GUGCGCAAACACAGGC--ACCACCCACC--------CGCCACCAACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUGAACUCAGUUCAGUUGGCCAAAAGCGAACCGCA ((((((.......(((--..........--------.)))......(((((((......)))))))(((((((((((((((....)))))))))))))))...)))...))). ( -47.50, z-score = -5.83, R) >droEre2.scaffold_4690 17928289 85 - 18748788 GUGCGCAAACACAG--------CCCACC--------CAU--CCGACGAGCCAACGCUUUUUGGUCCUGGGCCACUGAACUG----------AGUUGGCCAAAAGCGAACCGCA .((((........(--------((((..--------...--((((.((((....)))).))))...)))))..........----------.(((.((.....)).))))))) ( -23.80, z-score = -0.23, R) >droYak2.chrX 20585079 103 - 21770863 GUGCGCAAACACAGAACACCCAGCCACCCAAGCCAACACGGGCCAAGGGCCAACGCUUUUCGGUCCUGGCCAAGUGAACUG----------AGUUGGCCAAAAGCGAACCGCA (((.((................)))))....((((((...(((((.(((((..........)))))))))).((....)).----------.)))))).....(((...))). ( -32.79, z-score = -0.51, R) >droSec1.super_8 3732457 94 - 3762037 GUGCGCAAACACUGGCCAAACGACGGGC--------CACCACCGACGGCCCAAUG-CUUUUGGUCCUGGCCAACUGAACUG----------AGUUGGCCAAAAGCGAACCGCA ((((((.......(((((((.(..((((--------(.........)))))....-).))))))).(((((((((......----------)))))))))...)))...))). ( -38.00, z-score = -2.89, R) >droSim1.chrX 16531320 95 - 17042790 GUGCGCAAACACUGGCCAACCACCCACC--------CACCACCGACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUG----------AGUUGGCCAAAAGCGAACCGCA ((((((......(((....)))......--------..........(((((((......)))))))(((((((((......----------)))))))))...)))...))). ( -32.00, z-score = -1.78, R) >consensus GUGCGCAAACACAGGCCAACCACCCACC________CACCACCGACGGGCCAAUGCCUUUUGGUCCUGGCCAACUGAACUG__________AGUUGGCCAAAAGCGAACCGCA ((((((........................................(((((((......)))))))(((((((((................)))))))))...)))...))). (-22.59 = -23.79 + 1.20)

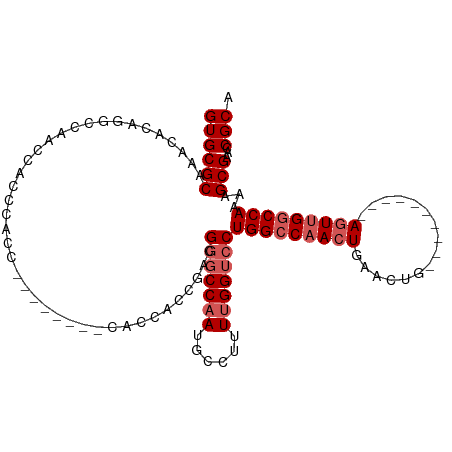
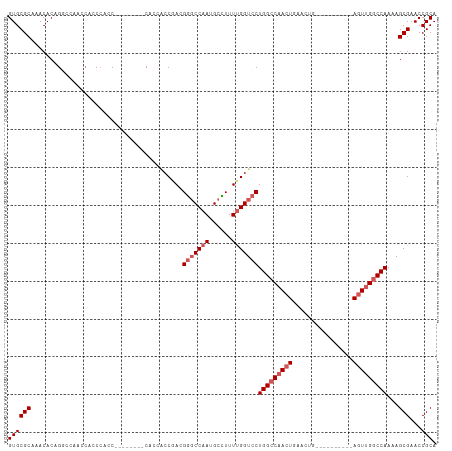
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:56 2011