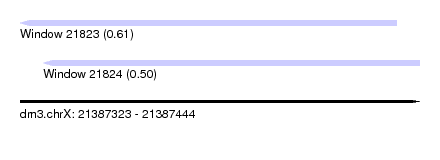
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,387,323 – 21,387,444 |
| Length | 121 |
| Max. P | 0.609931 |

| Location | 21,387,323 – 21,387,437 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Shannon entropy | 0.27049 |
| G+C content | 0.34622 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21387323 114 - 22422827 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))......................... ( -19.70, z-score = -1.23, R) >droSim1.chrX_random 5544770 114 - 5698898 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))......................... ( -19.70, z-score = -1.23, R) >droSec1.super_8 3705159 114 - 3762037 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))......................... ( -19.70, z-score = -1.23, R) >droYak2.chrX 20560290 114 - 21770863 AAGCCCUUAACGAAAAUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))......................... ( -20.20, z-score = -1.42, R) >droEre2.scaffold_4690 17905535 114 - 18748788 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))......................... ( -19.70, z-score = -1.23, R) >droAna3.scaffold_13117 2776062 114 + 5790199 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUUC .........(((.......))).(((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))).. ( -19.60, z-score = -0.93, R) >dp4.chrXL_group1a 7894722 114 + 9151740 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .........(((.......))).(((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))).. ( -19.60, z-score = -0.97, R) >droPer1.super_53 521121 114 - 525408 AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC .........(((.......))).(((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))).. ( -19.60, z-score = -0.97, R) >droWil1.scaffold_181096 11003850 106 + 12416693 AGGAAAAUGCCGAAAAUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUU-------- .((((((((.(((...(((.....)))(((((((..((.((....))..))..))))))).))).))....(((((..(((....)))..))))))))))).....-------- ( -19.20, z-score = -0.44, R) >droMoj3.scaffold_6359 3579252 99 + 4525533 ---------------CUGCUGCUAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCCAGGGUUCAAUUGUUUUUCACUUUUAUGCCUA ---------------.........((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))... ( -18.50, z-score = -1.27, R) >droGri2.scaffold_14853 6163615 99 + 10151454 ---------------CUGCCGCCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCCCG ---------------.........((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))... ( -18.50, z-score = -1.55, R) >droVir3.scaffold_13042 636051 99 + 5191987 --------CCUGGUGCCAGCAGCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGCUCGAUUGUUUUUCACCCCA------- --------.(((.((....)).)))..(((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).))....------- ( -22.40, z-score = -1.27, R) >consensus AAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUUAUGCUCC ........................((((((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).....)))... (-16.83 = -17.18 + 0.35)



| Location | 21,387,330 – 21,387,444 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Shannon entropy | 0.28913 |
| G+C content | 0.34755 |
| Mean single sequence MFE | -20.21 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 21387330 114 - 22422827 GCAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUU ((((....(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))...))))........... ( -20.50, z-score = -1.16, R) >droSim1.chrX_random 5544777 114 - 5698898 GCAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUU ((((....(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))...))))........... ( -20.50, z-score = -1.16, R) >droSec1.super_8 3705166 114 - 3762037 GCAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUU ((((....(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))...))))........... ( -20.50, z-score = -1.16, R) >droYak2.chrX 20560297 114 - 21770863 GCAGCCUAAGCCCUUAACGAAAAUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUU ((((....(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))...))))........... ( -21.00, z-score = -1.31, R) >droEre2.scaffold_4690 17905542 114 - 18748788 GCAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUCGCCCAAGGGUUCAAUUGUUUUUCACUUUU ((((....(((((((..(((((.(((.....)))(((((((..((.((....))..))..)))))))..............)))))...)))))))...))))........... ( -20.50, z-score = -1.16, R) >droAna3.scaffold_13117 2776069 114 + 5790199 GGAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUU (((((.(((((..........(((((.....)))))..........))))).))))).............((.((((.(((((..(((....)))..))))).)))).)).... ( -22.15, z-score = -1.02, R) >dp4.chrXL_group1a 7894729 114 + 9151740 UCGCUCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUU ......(((((..........(((((.....)))))..........)))))...................((.((((.(((((..(((....)))..))))).)))).)).... ( -18.35, z-score = -0.66, R) >droPer1.super_53 521128 114 - 525408 UCGCUCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUU ......(((((..........(((((.....)))))..........)))))...................((.((((.(((((..(((....)))..))))).)))).)).... ( -18.35, z-score = -0.66, R) >droWil1.scaffold_181096 11003850 113 + 12416693 GUAGUAUAGGAAAAUGCCGAAAAUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUU- ........((((((((.(((...(((.....)))(((((((..((.((....))..))..))))))).))).))....(((((..(((....)))..))))))))))).....- ( -19.20, z-score = -0.11, R) >droMoj3.scaffold_6359 3579259 94 + 4525533 --------------------UGCUGCUGCUAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCCAGGGUUCAAUUGUUUUUCACUUUU --------------------(((((....)))))(((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).... ( -19.50, z-score = -1.58, R) >droGri2.scaffold_14853 6163622 93 + 10151454 ---------------------GCUGCCGCCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUU ---------------------((((....)))).(((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).... ( -20.40, z-score = -2.31, R) >droVir3.scaffold_13042 636055 96 + 5191987 --------------UCCUGGUGCCAGCAGCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGCUCGAUUGUUUUUCAC---- --------------..(((.((....)).)))..(((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).))---- ( -21.60, z-score = -1.21, R) >consensus GCAGCCUAAGCCCUUAACGAAACUGCCGUCAGCAGUAAAUAUUUAUGCUUGUGUUUUAUUUAUUUACAUUGUCAAAACCAAUUUUGCCCAAGGGUUCAAUUGUUUUUCACUUUU .......................(((.....)))(((((((..((.((....))..))..)))))))...((.((((.(((((..(((....)))..))))).)))).)).... (-16.37 = -16.47 + 0.10)
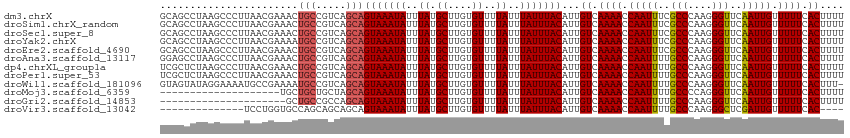


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:49 2011