
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,368,390 – 21,368,448 |
| Length | 58 |
| Max. P | 0.750641 |

| Location | 21,368,390 – 21,368,448 |
|---|---|
| Length | 58 |
| Sequences | 11 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Shannon entropy | 0.33086 |
| G+C content | 0.36156 |
| Mean single sequence MFE | -12.86 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.91 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21368390 58 - 22422827 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUG--------------- (((((.((((..((.(((.....))).))..)))))))))..((((.....))))...--------------- ( -13.20, z-score = -1.64, R) >droEre2.scaffold_4690 17888528 58 - 18748788 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGCGUGUG--------------- (((((.((((..((.(((.....))).))..)))))))))..................--------------- ( -9.60, z-score = -0.57, R) >droYak2.chrX 20542111 58 - 21770863 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUG--------------- (((((.((((..((.(((.....))).))..)))))))))..((((.....))))...--------------- ( -13.20, z-score = -1.64, R) >droSec1.super_8 3681230 58 - 3762037 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUG--------------- (((((.((((..((.(((.....))).))..)))))))))..((((.....))))...--------------- ( -13.20, z-score = -1.64, R) >droSim1.chrX 16501167 58 - 17042790 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGAGUGUG--------------- (((((.((((..((.(((.....))).))..)))))))))..((((.....))))...--------------- ( -13.20, z-score = -1.64, R) >dp4.chrXL_group1a 7872592 73 + 9151740 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGUGUGGACGAGGUCUGUGUAUG (((((.((((..((.(((.....))).))..))))))))).........(((..(((((...)))))..))). ( -15.20, z-score = -1.01, R) >droPer1.super_53 499025 73 - 525408 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUUUGUGUGUGUGGACGAGGUCUGUGUAUG (((((.((((..((.(((.....))).))..))))))))).........(((..(((((...)))))..))). ( -15.20, z-score = -1.01, R) >droAna3.scaffold_13117 2758592 58 + 5790199 AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACAC-ACACGGCCACAG-------------- (((((.((((..((.(((.....))).))..)))))))))......-............-------------- ( -9.60, z-score = -1.56, R) >droVir3.scaffold_13042 616791 58 + 5191987 AGGGAUGAUGGGAAUUUUGCUAAGAAUUUUUCAUUUCCUUUUACUCAGCGUGUGUAUG--------------- (((((.((((..((.(((.....))).))..))))))))).(((.(.....).)))..--------------- ( -10.20, z-score = -0.37, R) >droGri2.scaffold_14853 6146756 57 + 10151454 AGGGAUGAUGGGAAUUUCGUUAAGAAUUUUUCAUUUCCUUUAACGCAGCGUGUGUGU---------------- (((((.((((..((.(((.....))).))..)))))))))..((((.....))))..---------------- ( -15.20, z-score = -2.28, R) >droMoj3.scaffold_6359 3558909 57 + 4525533 AGGGAUGAUGGGAAUUUUGCUAAGAAUUUUUCAUUUCCUUUUACGCUGCGUGUGUGU---------------- (((((.((((..((.(((.....))).))..))))))))).(((((.....))))).---------------- ( -13.70, z-score = -1.72, R) >consensus AGGGAUGAUGGGAAUUUUACUAAGAAUUUUUCAUUUCCUUUAACUCUGUGUGUGUGUG_______________ (((((.((((..((.(((.....))).))..)))))))))................................. ( -9.99 = -9.91 + -0.08)

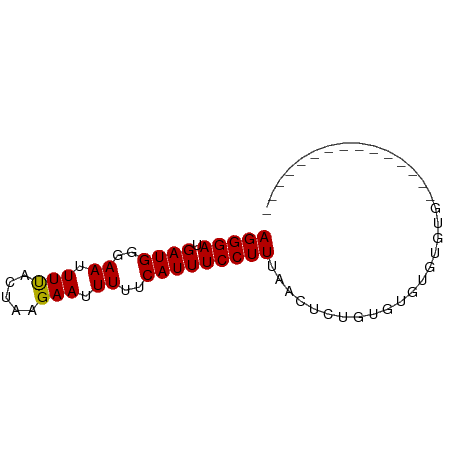
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:46 2011