| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,365,949 – 21,366,039 |
| Length | 90 |
| Max. P | 0.704546 |

| Location | 21,365,949 – 21,366,039 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.61638 |
| G+C content | 0.48226 |
| Mean single sequence MFE | -17.88 |
| Consensus MFE | -7.97 |
| Energy contribution | -7.93 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21365949 90 + 22422827 ----CAGCCUUUCCAUUCCCUUAGGGAUUUUCCCAC-UCCCAGCCGCUGCGUUUCAGUCAGCCAA-C----UUGGCUGUUAUAAAGUAAUUUUCGCCACU ----(((((..............(((.....)))..-........(((((......).))))...-.----..)))))...................... ( -18.20, z-score = -1.33, R) >droPer1.super_53 29095 89 - 525408 ----CCUCCUCUCUGUUCCU----AUGCUCCCUUGC-CCUGCUCUUUCGUUUGAACGUUAGCCAAAUU--CUGGGCUGUUAUAAAGUAAUUUUCGGCAUU ----................----.........(((-(.........(((....))).(((((.....--...)))))................)))).. ( -11.50, z-score = 0.57, R) >dp4.chrXL_group1a 1281796 89 + 9151740 ----CCUCCUCUCUGUUCCU----AUGCUCCCUUGC-CCUGCUCUUUCGUUUGAACGUUAGCCAAAUU--CUGGGCUGUUAUAAAGUAAUUUUCGGCAUU ----................----.........(((-(.........(((....))).(((((.....--...)))))................)))).. ( -11.50, z-score = 0.57, R) >droAna3.scaffold_13117 2757106 99 - 5790199 CACCCUGUUGCACCCCUUUUCAACGCGUUUUACC-CGCCACAUCUGUACUUUGUUAGUCAGCCAAAUUUACUUGGCUGUUAUAAAGUAAUUUUCGACACU .....(((((..............(((.......-)))........((((((((....(((((((......)))))))..)))))))).....))))).. ( -22.00, z-score = -3.48, R) >droEre2.scaffold_4690 17884999 87 + 18748788 ----CAGCC---CCAGUUCC--AGGACUUUUCCCGCAUCUCAGCCGCUGCUG---AGUCAGUCAA-CCAACUUGGCUGUUAUAAAGUAAUUUUCGCCACU ----(((((---..((((..--..((((..........((((((....))))---))..))))..-..)))).)))))...................... ( -20.10, z-score = -1.65, R) >droYak2.chrX 20539368 89 + 21770863 ----CAGCC---CCAUUUCC--AGGACUUUUCCCGC-UCUCCGCCGCUGCUUUUCAGUCAGCCAA-CUUGGUUGGCUGUUAUAAAGUAAUUUUCACCACU ----.(((.---........--.(((....))).((-.....)).)))(((((.(((((((((..-...)))))))))....)))))............. ( -21.20, z-score = -2.31, R) >droSec1.super_8 3678809 87 + 3762037 ----CAGCC---CCAUUCCCCCAGGGAUUUUGCAGC-UCUCAGCCGCUGCGUUUCAGUCAGCCAA-C----UUGGCUGUUAUAAAGUAAUUUUCGCCACU ----..(((---((.........)))((((((((((-........)))))........(((((..-.----..)))))....))))).......)).... ( -20.50, z-score = -0.88, R) >droSim1.chrX 16498735 87 + 17042790 ----CAGCC---CCAUUCCCCCAGGGAUUUUCCCGC-UCUCAGCCGCUGCGUUUCAGUCAGCCAA-C----UUGGCUGUUAUAAAGUAAUUUUCGCCACU ----(((((---...........(((.....)))((-.....)).(((((......).))))...-.----..)))))...................... ( -18.00, z-score = -0.80, R) >consensus ____CAGCC___CCAUUCCC__AGGGAUUUUCCCGC_UCUCAGCCGCUGCUUUUCAGUCAGCCAA_CU__CUUGGCUGUUAUAAAGUAAUUUUCGCCACU ..........................................................(((((((......)))))))...................... ( -7.97 = -7.93 + -0.05)

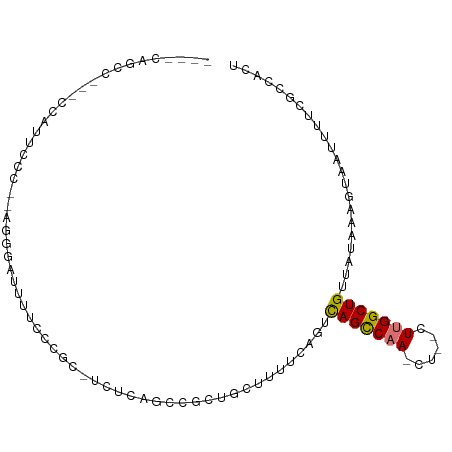
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:45 2011