| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,363,628 – 21,363,766 |
| Length | 138 |
| Max. P | 0.836226 |

| Location | 21,363,628 – 21,363,726 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.47821 |
| G+C content | 0.44557 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -12.78 |
| Energy contribution | -12.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
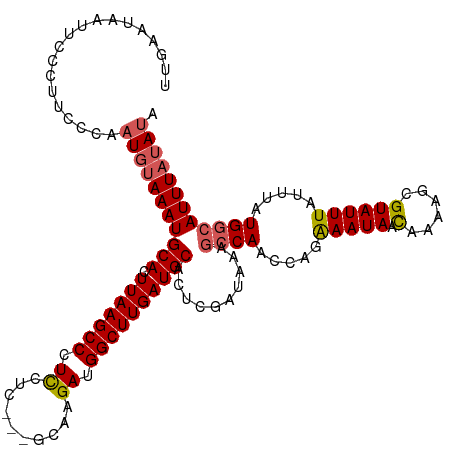
>dm3.chrX 21363628 98 + 22422827 -------------------CAUAAGUAAGUGGAUAAGCAAUUGAAUAAUUCCCUUCCCAAUGCAAAUGCACUUAAGCCCUUCUC---GCAAGAUGGCUUGAUGCACUCGAUAAACCCAAC -------------------..........(((....(((.(((..............))))))...((((.(((((((....((---....)).)))))))))))..........))).. ( -20.24, z-score = -1.62, R) >droSim1.chrX 16497591 101 + 17042790 -------------------AUUUUGCCAAGUCCCAAGCAAUUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUCCUCGCAAGAUGGCUUGAUGCACUCGAUAAAGCCAAC -------------------.....((...(((.(((....))).......................((((.(((((((.......((....)).)))))))))))...)))...)).... ( -16.40, z-score = -0.84, R) >droSec1.super_8 3676583 96 + 3762037 -------------------AUUUUG-----UCCCAAGCAAUUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUCCUCGCAAGAUGGCUUGAUGCACUCGAUAAAGCCAAC -------------------.(((((-----((.(((....))).......................((((.(((((((.......((....)).)))))))))))...)))))))..... ( -18.60, z-score = -2.17, R) >droYak2.chrX 20536898 117 + 21770863 GAGAGCUGAAAAGUUGAUGUCCGGAAGUAGUCGCAAUCAAUUGAAUAAUCCGCCUCCCGAUGUAAAUGCACUUAAGCCCUCCUG---GCUAGAUGGCUUGAUGCACUCAAUAAAGCCAAC (((((((....)))).(((((.(((.(..(...(((....)))......)..).))).)))))...((((.(((((((.((...---....)).))))))))))))))............ ( -26.70, z-score = -0.78, R) >droEre2.scaffold_4690 17882812 114 + 18748788 ---AGUUGGACAGUCGGGGUCACGAAAUAUCCCCAAUCAAUAGAACAAUACCCUUUCCAAUUUAAAUGCACUUAAGCCCUCCGC---GCGAGAUGGCUUGAUGCACUCGAUAAAGCCAAC ---.(((((...(((((((...........))))................................((((.(((((((.(((..---..).)).)))))))))))...)))....))))) ( -28.80, z-score = -1.91, R) >consensus ___________________ACUUGGAAAAGUCCCAAGCAAUUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUC___GCAAGAUGGCUUGAUGCACUCGAUAAAGCCAAC ..................................................................((((.(((((((.((..........)).)))))))))))............... (-12.78 = -12.62 + -0.16)


| Location | 21,363,649 – 21,363,766 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.27 |
| Shannon entropy | 0.15541 |
| G+C content | 0.37632 |
| Mean single sequence MFE | -22.41 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21363649 117 + 22422827 UUGAAUAAUUCCCUUCCCAAUGCAAAUGCACUUAAGCCCUUCUC---GCAAGAUGGCUUGAUGCACUCGAUAAACCCAACCAGAAAUAACAAAAGCGUAUUUAUUUAUGACAUUUAUAUA ...................((((...((((.(((((((....((---....)).))))))))))).((..............))..........))))...................... ( -17.34, z-score = -1.14, R) >droSim1.chrX 16497612 120 + 17042790 UUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUCCUCGCAAGAUGGCUUGAUGCACUCGAUAAAGCCAACCAGAAAUAACAAAAGCGUAUUUAUUUAUGGCAUUUAUAUA ...................(((((((((((.(((((((.......((....)).))))))))))..........((((.....(((((.(......)))))).....)))))))))))). ( -23.20, z-score = -1.91, R) >droSec1.super_8 3676599 120 + 3762037 UUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUCCUCGCAAGAUGGCUUGAUGCACUCGAUAAAGCCAACCAGAAAUAACAAAAGCGUAUUUAUUUAUGGCAUUUAUAUA ...................(((((((((((.(((((((.......((....)).))))))))))..........((((.....(((((.(......)))))).....)))))))))))). ( -23.20, z-score = -1.91, R) >droYak2.chrX 20536938 117 + 21770863 UUGAAUAAUCCGCCUCCCGAUGUAAAUGCACUUAAGCCCUCCUG---GCUAGAUGGCUUGAUGCACUCAAUAAAGCCAACCAGAAAUAAUAAAAGCGUAUUUAUUUAUGGCAUUUAUAUA ...........(((.......((((((((.(((.((((.....)---)))...((((((.((.......)).))))))..............))).))))))))....)))......... ( -25.50, z-score = -1.62, R) >droEre2.scaffold_4690 17882849 116 + 18748788 UAGAACAAUACCCUUUCCAAUUUAAAUGCACUUAAGCCCUCCGC---GCGAGAUGGCUUGAUGCACUCGAUAAAGCCAACCAGGAAUAACAAA-GCGUAUUUAUUUAUGGCAUUUAUACA ..........................((((.(((((((.(((..---..).)).)))))))))))....(((((((((.....(((((.....-...))))).....)))).)))))... ( -22.80, z-score = -1.26, R) >consensus UUGAAUAAUUCCCUUCCCAAUGUAAAUGCACUUAAGCCCUCCUC___GCAAGAUGGCUUGAUGCACUCGAUAAAGCCAACCAGAAAUAACAAAAGCGUAUUUAUUUAUGGCAUUUAUAUA ...................(((((((((((.(((((((.((..........)).))))))))))..........((((.....(((((.(......)))))).....)))))))))))). (-18.54 = -19.06 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:44 2011