| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,338,923 – 21,339,029 |
| Length | 106 |
| Max. P | 0.707004 |

| Location | 21,338,923 – 21,339,026 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.01 |
| Shannon entropy | 0.39981 |
| G+C content | 0.38982 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.19 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21338923 103 + 22422827 ----------GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCC----GAAAGC-UUCGGGUGUCCUG- ----------..........((((((.((((.(((.....))))))).)))))).((....((((((((......)))))))).))((..((((----((....-.))))).)..)).- ( -29.10, z-score = -2.28, R) >droSim1.chrX 16486523 103 + 17042790 ----------GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCC----GAAAGC-UCCGGGUGUCCUG- ----------..........((((((.((((.(((.....))))))).))))))..((((.((((((((......)))))))).((.((((...----....))-))))...))))..- ( -28.50, z-score = -2.07, R) >droSec1.super_8 3651535 103 + 3762037 ----------GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCC----GAAAGC-UCCGGGUGUCCUG- ----------..........((((((.((((.(((.....))))))).))))))..((((.((((((((......)))))))).((.((((...----....))-))))...))))..- ( -28.50, z-score = -2.07, R) >droYak2.chrX 20512704 104 + 21770863 ----------GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCCC---CAAAGC-UCCGGGUGUCCUG- ----------..........((((((.((((.(((.....))))))).))))))..((((.((((((((......)))))))).((.((((....---....))-))))...))))..- ( -28.80, z-score = -2.07, R) >droEre2.scaffold_4690 17859414 102 + 18748788 ----------GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCC-----GAAAGC-UCCGGGUGUCCUG- ----------..........((((((.((((.(((.....))))))).))))))..((((.((((((((......))))))))......((((-----(.....-..)))))))))..- ( -29.40, z-score = -2.48, R) >droAna3.scaffold_13117 2734185 109 - 5790199 ----------GAAAAAAUGAUGCCGUAAAGUUGAUUAGAAGUCACUUAAUAGCAGCGGAUGUUCAGGUUUAUUACAAUUUGUAACGAGAGCCCCUCUGAAAAGCACUCCGGGGUUUCGU ----------(((....((.(((..(.((((.(((.....))))))).)..))).))....)))........(((.....)))((((((.((((.((....))......)))))))))) ( -22.30, z-score = 0.19, R) >dp4.chrXL_group1a 7806284 89 - 9151740 ----------AAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCUCUUCCAG-------------------- ----------..........((((((.((((.(((.....))))))).))))))..(((.(.((...((..((((.....))))..)).)).).)))..-------------------- ( -22.70, z-score = -2.28, R) >droPer1.super_53 453329 89 + 525408 ----------AAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCUCUUCCAG-------------------- ----------..........((((((.((((.(((.....))))))).))))))..(((.(.((...((..((((.....))))..)).)).).)))..-------------------- ( -22.70, z-score = -2.28, R) >droWil1.scaffold_181096 10947716 113 - 12416693 AAAAAAAAUGUACGAUGCGAUGCCAUAAAGUUUAUUAGAAGUCACUUAAUAGCAACGGAUGUGCAAGUUUAUUACAAUUUGUAACGAGAGUGCUC---AAAA-CUGUUGACUUACAU-- .........((((.((.((.(((.((.((((............)))).)).))).)).))))))(((((....(((.((((..((....))...)---))).-.))).)))))....-- ( -19.30, z-score = 0.25, R) >droVir3.scaffold_13042 4484463 89 + 5191987 ----------GAAAAAAUGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUACGAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAAG----------------- ----------.......((.((((((.((((............)))).)))))).))....((((((((......))))))))..(((....)))---....----------------- ( -17.40, z-score = -1.24, R) >droMoj3.scaffold_6359 868990 104 + 4525533 ----------AAAAAAAAGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUGCAAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAGAGCUGUUGAUUUACAU-- ----------........(.((((((.((((............)))).)))))).).....((((((((......)))))))).(((.(((((..---..))))).)))........-- ( -25.40, z-score = -2.26, R) >droGri2.scaffold_14853 3934957 104 + 10151454 ----------AAAAAAAUGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUGCGAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAAAGCUGUUGAUUUACAU-- ----------...........(((((.((((............)))).)))))(((((...((.((((((.((((.....))))...)))))).)---)....))))).........-- ( -22.20, z-score = -1.10, R) >consensus __________GAAAAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCUC___GAAAGC_UUCGGGUGUCCU__ ....................((((((.((((............)))).)))))).((....((((((((......)))))))).))................................. (-14.63 = -14.19 + -0.44)
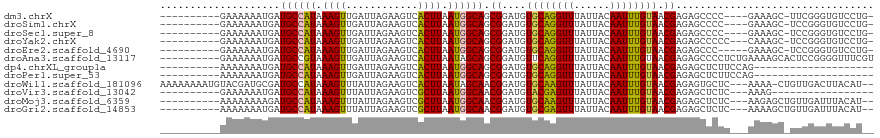
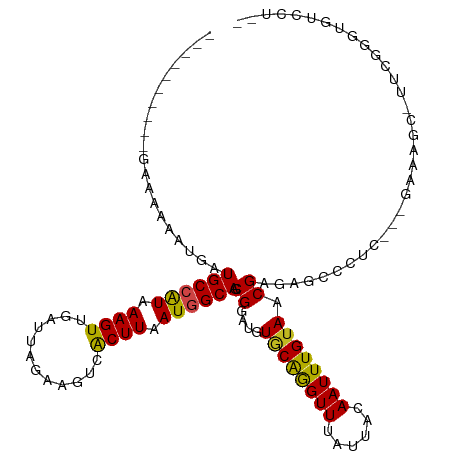

| Location | 21,338,923 – 21,339,026 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.01 |
| Shannon entropy | 0.39981 |
| G+C content | 0.38982 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -10.93 |
| Energy contribution | -10.87 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21338923 103 - 22422827 -CAGGACACCCGAA-GCUUUC----GGGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC---------- -((((...(((((.-....))----)))......(((((.....)))))...)))).......(.((((((.((((((.......)).)))).)))))).)........---------- ( -26.50, z-score = -3.13, R) >droSim1.chrX 16486523 103 - 17042790 -CAGGACACCCGGA-GCUUUC----GGGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC---------- -((((...((((((-...)))----)))......(((((.....)))))...)))).......(.((((((.((((((.......)).)))).)))))).)........---------- ( -26.00, z-score = -2.51, R) >droSec1.super_8 3651535 103 - 3762037 -CAGGACACCCGGA-GCUUUC----GGGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC---------- -((((...((((((-...)))----)))......(((((.....)))))...)))).......(.((((((.((((((.......)).)))).)))))).)........---------- ( -26.00, z-score = -2.51, R) >droYak2.chrX 20512704 104 - 21770863 -CAGGACACCCGGA-GCUUUG---GGGGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC---------- -((((......(((-((((..---..))))))).(((((.....)))))...)))).......(.((((((.((((((.......)).)))).)))))).)........---------- ( -26.10, z-score = -1.58, R) >droEre2.scaffold_4690 17859414 102 - 18748788 -CAGGACACCCGGA-GCUUUC-----GGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC---------- -((((......(((-(((...-----.)))))).(((((.....)))))...)))).......(.((((((.((((((.......)).)))).)))))).)........---------- ( -25.60, z-score = -2.78, R) >droAna3.scaffold_13117 2734185 109 + 5790199 ACGAAACCCCGGAGUGCUUUUCAGAGGGGCUCUCGUUACAAAUUGUAAUAAACCUGAACAUCCGCUGCUAUUAAGUGACUUCUAAUCAACUUUACGGCAUCAUUUUUUC---------- ..((..((((.(((.....)))...))))...))(((((.....)))))..............(.((((...((((((.......)).))))...)))).)........---------- ( -17.70, z-score = 0.66, R) >dp4.chrXL_group1a 7806284 89 + 9151740 --------------------CUGGAAGAGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUU---------- --------------------..(((.(.((....(((((.....)))))......)).).)))(.((((((.((((((.......)).)))).)))))).)........---------- ( -19.00, z-score = -2.35, R) >droPer1.super_53 453329 89 - 525408 --------------------CUGGAAGAGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUU---------- --------------------..(((.(.((....(((((.....)))))......)).).)))(.((((((.((((((.......)).)))).)))))).)........---------- ( -19.00, z-score = -2.35, R) >droWil1.scaffold_181096 10947716 113 + 12416693 --AUGUAAGUCAACAG-UUUU---GAGCACUCUCGUUACAAAUUGUAAUAAACUUGCACAUCCGUUGCUAUUAAGUGACUUCUAAUAAACUUUAUGGCAUCGCAUCGUACAUUUUUUUU --.(((((((..((((-(((.---.(((......)))..))))))).....)))))))....((.((((((.((((............)))).)))))).))................. ( -20.50, z-score = -1.17, R) >droVir3.scaffold_13042 4484463 89 - 5191987 -----------------CUUU---GAGAGCUCUCGUUACAAAUUGUAAUAAACUCGUACAUCCGUUGCCAUUAAGCGACUUCUAAUAAACUUUAUGGCAUCAUUUUUUC---------- -----------------....---(((....)))(((((.....)))))..............(.((((((.(((..............))).)))))).)........---------- ( -13.64, z-score = -1.15, R) >droMoj3.scaffold_6359 868990 104 - 4525533 --AUGUAAAUCAACAGCUCUU---GAGAGCUCUCGUUACAAAUUGUAAUAAACUUGCACAUCCGUUGCCAUUAAGCGACUUCUAAUAAACUUUAUGGCAUCUUUUUUUU---------- --.(((((......(((((..---..)))))...(((((.....)))))....))))).....(.((((((.(((..............))).)))))).)........---------- ( -19.04, z-score = -1.66, R) >droGri2.scaffold_14853 3934957 104 - 10151454 --AUGUAAAUCAACAGCUUUU---GAGAGCUCUCGUUACAAAUUGUAAUAAACUCGCACAUCCGUUGCCAUUAAGCGACUUCUAAUAAACUUUAUGGCAUCAUUUUUUU---------- --.............((....---(((....)))(((((.....)))))......))......(.((((((.(((..............))).)))))).)........---------- ( -15.84, z-score = -0.56, R) >consensus __AGGACACCCGAA_GCUUUC___GAGGGCUCUCGUUACAAAUUGUAAUAAACCUGCACAUCCGCUGCCAUUAAGUGACUUCUAAUCAACUUUAUGGCAUCAUUUUUUC__________ ..................................(((((.....)))))..............(.((((((.(((..............))).)))))).).................. (-10.93 = -10.87 + -0.05)



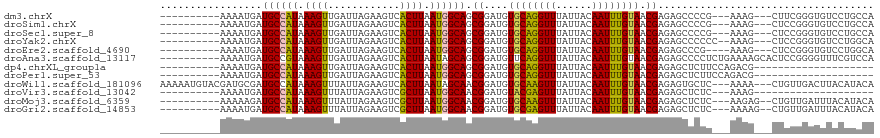
| Location | 21,338,926 – 21,339,029 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Shannon entropy | 0.39247 |
| G+C content | 0.40076 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.19 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 21338926 103 + 22422827 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCCG---AAAG---CUUCGGGUGUCCUGCCA ----------....((.((((((.((((.(((.....))))))).)))))).))..((.(((((((..((((.....))))..)).((((((---(...---..))))).))))))))) ( -31.90, z-score = -2.53, R) >droSim1.chrX 16486526 103 + 17042790 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCCG---AAAG---CUCCGGGUGUCCUGCCA ----------....((.((((((.((((.(((.....))))))).)))))).))..((.(((((((..((((.....))))..)).((((.(---(...---.)).))))..))))))) ( -30.40, z-score = -2.03, R) >droSec1.super_8 3651538 103 + 3762037 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCCG---AAAG---CUCCGGGUGUCCUGCCA ----------....((.((((((.((((.(((.....))))))).)))))).))..((.(((((((..((((.....))))..)).((((.(---(...---.)).))))..))))))) ( -30.40, z-score = -2.03, R) >droYak2.chrX 20512707 104 + 21770863 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCCCC--AAAG---CUCCGGGUGUCCUGGCA ----------.......((((((.((((.(((.....))))))).))))))((((((.((((((((......)))))))).((.((((.....--...)---)))))...))))..)). ( -29.80, z-score = -1.40, R) >droEre2.scaffold_4690 17859417 102 + 18748788 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCG----AAAG---CUCCGGGUGUCCUGGCA ----------.......((((((.((((.(((.....))))))).))))))((((((.((((((((......))))))))......(((((----....---...)))))))))..)). ( -30.40, z-score = -1.89, R) >droAna3.scaffold_13117 2734188 109 - 5790199 ----------AAAAUGAUGCCGUAAAGUUGAUUAGAAGUCACUUAAUAGCAGCGGAUGUUCAGGUUUAUUACAAUUUGUAACGAGAGCCCCUCUGAAAAGCACUCCGGGGUUUCGUCCA ----------.......(((..(.((((.(((.....))))))).)..)))..(((((((((((((......)))))).)))((((.((((.((....))......)))))))))))). ( -25.10, z-score = -0.48, R) >dp4.chrXL_group1a 7806287 89 - 9151740 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCUCUUCCAGACG-------------------- ----------.......((((((.((((.(((.....))))))).))))))..(((.(.((...((..((((.....))))..)).)).).))).....-------------------- ( -22.70, z-score = -1.92, R) >droPer1.super_53 453332 89 + 525408 ----------AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCUCUUCCAGACG-------------------- ----------.......((((((.((((.(((.....))))))).))))))..(((.(.((...((..((((.....))))..)).)).).))).....-------------------- ( -22.70, z-score = -1.92, R) >droWil1.scaffold_181096 10947719 113 - 12416693 AAAAAUGUACGAUGCGAUGCCAUAAAGUUUAUUAGAAGUCACUUAAUAGCAACGGAUGUGCAAGUUUAUUACAAUUUGUAACGAGAGUGCUC---AAAA---CUGUUGACUUACAUACA ....(((((.(....(((....(((......)))...))).........((((((.((.(((..((..((((.....))))..))..))).)---)...---)))))).).)))))... ( -19.80, z-score = 0.44, R) >droVir3.scaffold_13042 4484466 86 + 5191987 ----------AAAAUGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUACGAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAAG-------------------- ----------....((.((((((.((((............)))).)))))).))....((((((((......))))))))..(((....)))---....-------------------- ( -17.40, z-score = -1.25, R) >droMoj3.scaffold_6359 868993 104 + 4525533 ----------AAAAAGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUGCAAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAGAG--CUGUUGAUUUACAUACA ----------.....(.((((((.((((............)))).)))))).)(.(((((((((((......)))))))..(((.(((((..---..)))--)).)))....)))).). ( -26.40, z-score = -2.33, R) >droGri2.scaffold_14853 3934960 104 + 10151454 ----------AAAAUGAUGCCAUAAAGUUUAUUAGAAGUCGCUUAAUGGCAACGGAUGUGCGAGUUUAUUACAAUUUGUAACGAGAGCUCUC---AAAAG--CUGUUGAUUUACAUACA ----------...(((..(((((.((((............)))).)))))(((((...((.((((((.((((.....))))...)))))).)---)....--)))))......)))... ( -22.90, z-score = -1.06, R) >consensus __________AAAAUGAUGCCAUAAAGUUGAUUAGAAGUCACUUAAUGGCAGCGGAUGUGCAGGUUUAUUACAAUUUGUAACGAGAGCCCUC___AAAG___CUCCGGGUGUCCUGCCA .................((((((.((((............)))).)))))).((....((((((((......)))))))).)).................................... (-14.63 = -14.19 + -0.44)
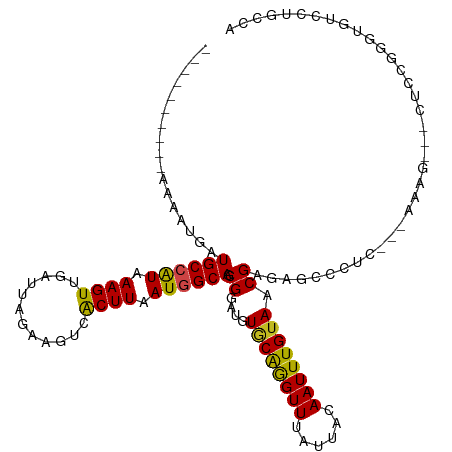
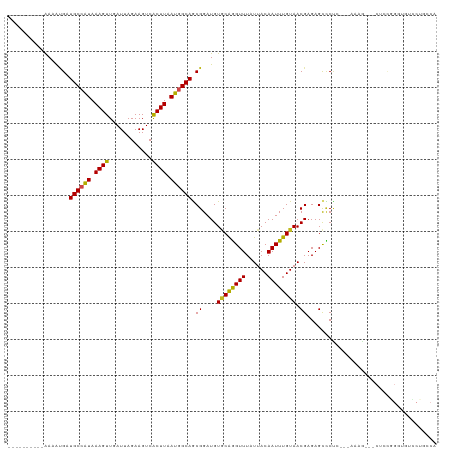
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:38 2011