| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,332,080 – 21,332,132 |
| Length | 52 |
| Max. P | 0.687045 |

| Location | 21,332,080 – 21,332,132 |
|---|---|
| Length | 52 |
| Sequences | 4 |
| Columns | 54 |
| Reading direction | forward |
| Mean pairwise identity | 50.94 |
| Shannon entropy | 0.80869 |
| G+C content | 0.37019 |
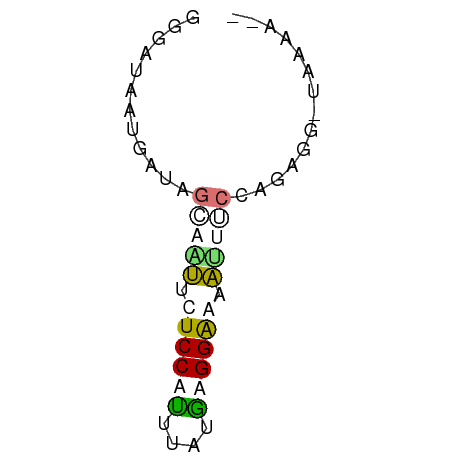
| Mean single sequence MFE | -8.74 |
| Consensus MFE | -4.19 |
| Energy contribution | -5.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.30 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687045 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
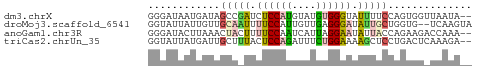
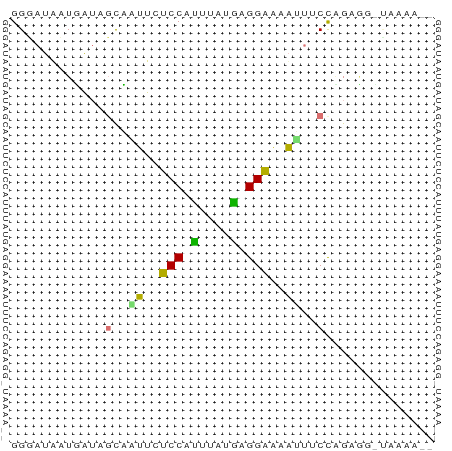
>dm3.chrX 21332080 52 + 22422827 GGGAUAAUGAUAGCCGAUCUCCAUGUAUGUGGGUAUUUUCCAGUGGUUAAUA-- .(((....((((.(((..(.....)....))).)))).)))...........-- ( -8.00, z-score = 1.10, R) >droMoj3.scaffold_6541 413312 52 - 2543558 GGUAUUAUUGUUGCAAUUUUCCAUUGUUGAGGGAUAUUGCUGGUG--UCAAGUA (..((((.....(((((.((((........)))).))))))))).--.)..... ( -10.30, z-score = -0.78, R) >anoGam1.chr3R 34125638 52 - 53272125 GGGAUACUUAAACUACUUUUCCAAUCAUUAGGAAUAUUACCAGAAGACCAAA-- ((....(((.........((((........)))).........))).))...-- ( -5.57, z-score = -0.23, R) >triCas2.chrUn_35 169544 52 + 346786 GGUAUUAUGAUUGCUUUACUCCAGAUUUCUGGAAAAGCUCCUGACUCAAAGA-- ............(((((..(((((....))))))))))..............-- ( -11.10, z-score = -1.28, R) >consensus GGGAUAAUGAUAGCAAUUCUCCAUUUAUGAGGAAAAUUUCCAGAGG_UAAAA__ ............(((((.((((((....)))))).))))).............. ( -4.19 = -5.00 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:36 2011