| Sequence ID | dm3.chrX |
|---|---|
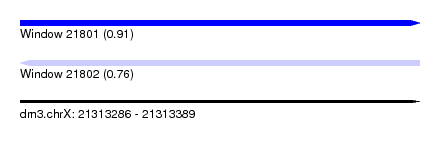
| Location | 21,313,286 – 21,313,389 |
| Length | 103 |
| Max. P | 0.911128 |

| Location | 21,313,286 – 21,313,389 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 55.17 |
| Shannon entropy | 0.89237 |
| G+C content | 0.35031 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -3.14 |
| Energy contribution | -4.54 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.80 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.14 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
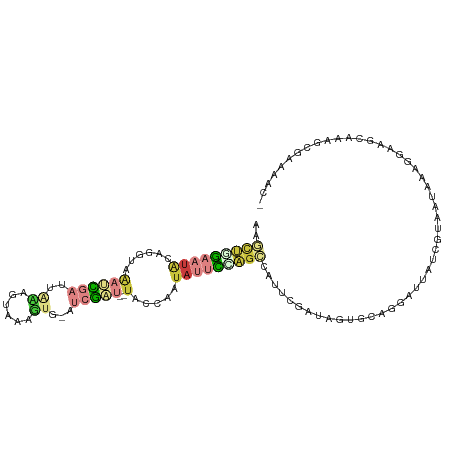
>dm3.chrX 21313286 103 + 22422827 AAGCUGGAAUACAGGUAAAUCGAUUAAAGUCAAGUG-GUCGAU--UACC-AUAUUCUAGCCAUUCGAUAGUGCAGGAUUAUCGUAAUAACGGAAGCAAAGCGAAAAC- ..(((((((((..(((((.(((((((........))-))))))--))))-.)))))))))..((((....(((.......((((....))))..)))...))))...- ( -28.20, z-score = -3.63, R) >droSec1.super_8 3632365 104 + 3762037 AAGCUGGAAUACAGAUAAAUCGAUUACAGUAAAGUG-CUCGAU--UACCCAUAUUCCAGCGAUUCGAUAGUGCAGGAUUAUCGUAAUAAAGGAAGCAAAGCGAAAAC- ..(((((((((......((((((.(((......)))-.)))))--).....))))))))).((((((((((.....))))))).)))....................- ( -26.30, z-score = -3.28, R) >droYak2.chrX 20491757 104 + 21770863 AAGCCGGAAUGUAGGUAAAUCGAUUACUGUAGAGUU-GUCGAU--UACCAAUAUUCCAGCCAUUCGAUAGUGCAGGAUUAUCGUAAUAAAGGAAGCAAAGCGAAAAC- ..((.(((((((.(((((.(((((.(((....))).-))))))--)))).)))))))..((((((((((((.....))))))).)))...))..))...........- ( -29.80, z-score = -3.99, R) >droEre2.scaffold_4690 17840554 103 + 18748788 AAGCUGGAAUAUAUGUAAAUCGAUUACAGU-GAGUG-AUCGAU--UACCAAUAUUCGAGCCAUUCGACAGUGCAGGAUUAUCGUAAUAAAGGAAGCAAAGCGAAAAC- ..(((.((((((.((..((((((((((...-..)))-))))))--)..)))))))).)))..((((.(..(((....((((....)))).....)))..)))))...- ( -27.60, z-score = -3.57, R) >droPer1.super_53 423928 87 + 525408 CCACUGGAGUACAAUUCAAUUGAC-GGGGUAACUUC-AUAAUG--AAUUGAGAAUUAAGUAAUUGUACACAAAUUGUUAACGAAAACAAAU----------------- ..(((.((...(((((((..(((.-(......).))-)...))--)))))....)).))).............(((((......)))))..----------------- ( -10.80, z-score = 0.54, R) >droWil1.scaffold_181096 10922637 99 - 12416693 AAAGCAAAAUAUAU-UCAAAUGUUUAAGAUAAUUACAAUAAUCGAUAUUGAUAUGUCGACAAAUCGAUUUGGUUAUCGUUACGUACUUAUUUGCCCAAAC-------- ...(((((((((..-....)))).((((.((...((.((((((((.((((((.((....)).)))))))))))))).))....)))))))))))......-------- ( -15.90, z-score = -0.85, R) >droGri2.scaffold_14853 3913357 99 + 10151454 UAUUUUGAAUGCAUAUCGAUUGCUUCAAUUCAAUAGCAUCGUU--UUCCCAU---UAAGUUAU-CGAUAACGUAAUAUU-UGCCGAUAACUCGAU--AGGCAAUCAUU .....(((.(((.((((((.((((..........)))).....--.......---...(((((-((.(((........)-)).))))))))))))--).))).))).. ( -22.00, z-score = -2.69, R) >consensus AAGCUGGAAUACAGGUAAAUCGAUUAAAGUAAAGUG_AUCGAU__UACCAAUAUUCCAGCCAUUCGAUAGUGCAGGAUUAUCGUAAUAAAGGAAGCAAAGCGAAAAC_ ..(((((((((.......((((((.............))))))........)))))))))................................................ ( -3.14 = -4.54 + 1.39)


| Location | 21,313,286 – 21,313,389 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 55.17 |
| Shannon entropy | 0.89237 |
| G+C content | 0.35031 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -1.71 |
| Energy contribution | -3.26 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.86 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.08 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
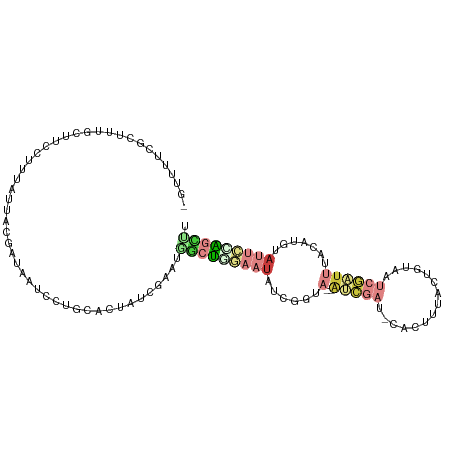
>dm3.chrX 21313286 103 - 22422827 -GUUUUCGCUUUGCUUCCGUUAUUACGAUAAUCCUGCACUAUCGAAUGGCUAGAAUAU-GGUA--AUCGAC-CACUUGACUUUAAUCGAUUUACCUGUAUUCCAGCUU -...((((...(((....(((((....)))))...)))....)))).((((.((((((-((((--(((((.-.............)))).))))).)))))).)))). ( -21.44, z-score = -2.12, R) >droSec1.super_8 3632365 104 - 3762037 -GUUUUCGCUUUGCUUCCUUUAUUACGAUAAUCCUGCACUAUCGAAUCGCUGGAAUAUGGGUA--AUCGAG-CACUUUACUGUAAUCGAUUUAUCUGUAUUCCAGCUU -...((((...(((.....((((....))))....)))....))))..(((((((((((((((--(((((.-.((......))..)))).)))))))))))))))).. ( -32.30, z-score = -5.38, R) >droYak2.chrX 20491757 104 - 21770863 -GUUUUCGCUUUGCUUCCUUUAUUACGAUAAUCCUGCACUAUCGAAUGGCUGGAAUAUUGGUA--AUCGAC-AACUCUACAGUAAUCGAUUUACCUACAUUCCGGCUU -...((((...(((.....((((....))))....)))....)))).(((((((((...((((--(((((.-.(((....)))..)))).)))))...))))))))). ( -27.10, z-score = -4.06, R) >droEre2.scaffold_4690 17840554 103 - 18748788 -GUUUUCGCUUUGCUUCCUUUAUUACGAUAAUCCUGCACUGUCGAAUGGCUCGAAUAUUGGUA--AUCGAU-CACUC-ACUGUAAUCGAUUUACAUAUAUUCCAGCUU -...(((((..(((.....((((....))))....)))..).)))).((((.((((((((..(--((((((-.((..-...)).)))))))..)).)))))).)))). ( -24.60, z-score = -3.44, R) >droPer1.super_53 423928 87 - 525408 -----------------AUUUGUUUUCGUUAACAAUUUGUGUACAAUUACUUAAUUCUCAAUU--CAUUAU-GAAGUUACCCC-GUCAAUUGAAUUGUACUCCAGUGG -----------------..(((((......)))))............((((.......(((((--((...(-((.(......)-.)))..)))))))......)))). ( -9.82, z-score = 0.19, R) >droWil1.scaffold_181096 10922637 99 + 12416693 --------GUUUGGGCAAAUAAGUACGUAACGAUAACCAAAUCGAUUUGUCGACAUAUCAAUAUCGAUUAUUGUAAUUAUCUUAAACAUUUGA-AUAUAUUUUGCUUU --------(((((((...((((.((((((((((((......((((....))))........))))).))).)))).)))))))))))......-.............. ( -14.44, z-score = -0.07, R) >droGri2.scaffold_14853 3913357 99 - 10151454 AAUGAUUGCCU--AUCGAGUUAUCGGCA-AAUAUUACGUUAUCG-AUAACUUA---AUGGGAA--AACGAUGCUAUUGAAUUGAAGCAAUCGAUAUGCAUUCAAAAUA ........(((--((..((((((((..(-(........))..))-))))))..---)))))..--...(((((((((((.(((...))))))))).)))))....... ( -26.10, z-score = -3.17, R) >consensus _GUUUUCGCUUUGCUUCCUUUAUUACGAUAAUCCUGCACUAUCGAAUGGCUGGAAUAUCGGUA__AUCGAU_CACUUUACUGUAAUCGAUUUACAUGUAUUCCAGCUU ...............................................(((((((((.........(((((...............)))))........))))))))). ( -1.71 = -3.26 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:31 2011