| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,288,077 – 21,288,183 |
| Length | 106 |
| Max. P | 0.963913 |

| Location | 21,288,077 – 21,288,183 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Shannon entropy | 0.23065 |
| G+C content | 0.37748 |
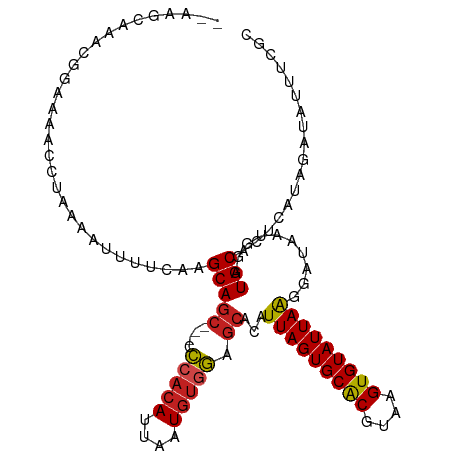
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
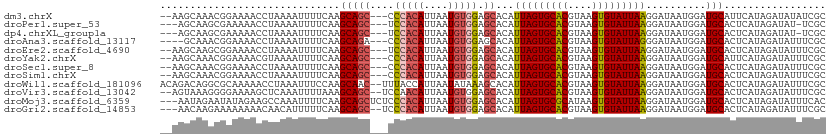
>dm3.chrX 21288077 106 - 22422827 --AAGCAAACGGAAAACCUAAAAUUUUCAAGCAGC---CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCAUUCAUAGAUAUAUCGC --..(((..((.....(((..............((---.(((((....))))).))....((((((((....)))))))))))....))..)))................. ( -27.10, z-score = -2.78, R) >droPer1.super_53 387182 104 - 525408 ---AGCAAGCGAAAAACCUAAAAUUUUCAAGCAGC---UCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAU-UCGC ---.....(((((...(((..............((---((((((....))))))))....((((((((....)))))))))))...((((......)))).....)-)))) ( -32.90, z-score = -4.54, R) >dp4.chrXL_group1a 7739233 104 + 9151740 ---AGCAAGCGAAAAACCUAAAAUUUUCAAGCAGC---UCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAU-UCGC ---.....(((((...(((..............((---((((((....))))))))....((((((((....)))))))))))...((((......)))).....)-)))) ( -32.90, z-score = -4.54, R) >droAna3.scaffold_13117 2690366 104 + 5790199 ----GCAAACGGAAAACCUAAAAUUUUCAAGCAGA---CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAGGGAUAAUGGAUGCACUCAUAGAUAUUUCGC ----(((..((.....(((((............(.---.(((((....)))))..).......(((((....)))))))))).....))..)))................. ( -24.30, z-score = -1.79, R) >droEre2.scaffold_4690 17816202 106 - 18748788 --AAGCAAGCGGAAAACCUAAAAUUUUCAAGCAGC---UCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC --......((((((..(((..............((---((((((....))))))))....((((((((....)))))))))))...((((......)))).....)))))) ( -33.30, z-score = -4.40, R) >droYak2.chrX 20465603 106 - 21770863 --AAGCAAACGGAAAACGUAAAAUUUUCAAGCAGC---CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCAUUCAUAGAUAUUUCGC --..(((.(((.....)))......((((....((---.(((((....))))).))...(((((((((....)))))))))......)))))))................. ( -27.10, z-score = -2.77, R) >droSec1.super_8 3603059 106 - 3762037 --AAGCAAACGGAAAACCUAAAAUUUUCAAGCAGC---CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC --..(((..((.....(((..............((---.(((((....))))).))....((((((((....)))))))))))....))..)))................. ( -27.10, z-score = -2.60, R) >droSim1.chrX 16428995 106 - 17042790 --AAGCAAACGGAAAACCUAAAAUUUUCAAGCAGC---CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC --..(((..((.....(((..............((---.(((((....))))).))....((((((((....)))))))))))....))..)))................. ( -27.10, z-score = -2.60, R) >droWil1.scaffold_181096 1522980 109 - 12416693 ACAGACAGGCGCAAAAACCUAAAUUUCCAAGCAAC--UUUACCAUUAAUAUAAAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC ........(((((....((......(((......(--((((.........)))))....(((((((((....))))))))))))....)).))).......((....)))) ( -18.60, z-score = -0.60, R) >droVir3.scaffold_13042 4394996 107 - 5191987 --AGUAAAGGGGAAAAGCUCAAAUUUUAAAGCAGC--UCCAACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC --......(((((.....))..........(((((--((((.((....))))))))...(((((((((....)))))))))..........))).)))............. ( -26.70, z-score = -2.44, R) >droMoj3.scaffold_6359 807126 108 - 4525533 ---AAUAGAAUAUAGAAGCCAAAUUUUCAAGCAGCUCUCCCACAUUAAUGUGGAGCACAUUAGUGCGCAUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCAC ---....((((((.((((......))))..((.......(((((....))))).((((....))))))...((((((((..........)))))))).....))))))... ( -22.10, z-score = -0.89, R) >droGri2.scaffold_14853 3887974 106 - 10151454 ---AACAAGAAAAAAAACAACAUUUUUCAAGCAGC--UCCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC ---.....((((((........))))))..(((((--(((.(((....))))))))...(((((((((....)))))))))..........)))................. ( -27.60, z-score = -3.57, R) >consensus __AAGCAAACGGAAAACCUAAAAUUUUCAAGCAGC___CCCACAUUAAUGUGGAGCACAUUAGUGCACGUAAGUGUAUUAAGGAUAAUGGAUGCACUCAUAGAUAUUUCGC ..............................(((......(((((....)))))......(((((((((....)))))))))..........)))................. (-18.70 = -18.73 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:28 2011