| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,267,245 – 21,267,314 |
| Length | 69 |
| Max. P | 0.500000 |

| Location | 21,267,245 – 21,267,314 |
|---|---|
| Length | 69 |
| Sequences | 11 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Shannon entropy | 0.47438 |
| G+C content | 0.34825 |
| Mean single sequence MFE | -9.37 |
| Consensus MFE | -3.50 |
| Energy contribution | -3.39 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
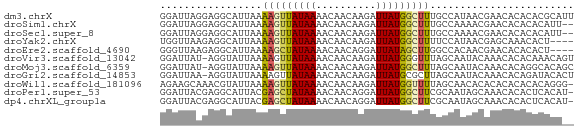
>dm3.chrX 21267245 69 - 22422827 GGAUUAGGAGGCAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUGCCAUAACGAACACACACGCAUU .........((((....(((((((((..........))))))))))))).................... ( -10.20, z-score = -1.84, R) >droSim1.chrX 16404882 67 - 17042790 GGAUUAGGAGGCAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUGCCAAAACGAACACACACAUU-- .........((((....(((((((((..........)))))))))))))..................-- ( -10.20, z-score = -2.55, R) >droSec1.super_8 3577313 67 - 3762037 GGAUUAGGAGGCAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUGCCAAAACGAACACACACAUU-- .........((((....(((((((((..........)))))))))))))..................-- ( -10.20, z-score = -2.55, R) >droYak2.chrX 20446394 65 - 21770863 UGGUUAAGAGGCAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUUCCAUAACGAGCAAACACU---- ..((((...((....(((((((((((..........))))))))))))).))))...........---- ( -7.80, z-score = -0.56, R) >droEre2.scaffold_4690 17797823 65 - 18748788 GGGUUAAGAGGCAUUAAAAGCUAUAAAACAACAGGAUUAUAGCUUGGCCACAACGAACACACACU---- .........(((.....(((((((((..(....)..))))))))).)))................---- ( -15.00, z-score = -4.29, R) >droVir3.scaffold_13042 4367064 68 - 5191987 GGAUUAU-AGGUAUUAAAAGUUAUAAAACAACAAGAUUAUGGGUUUAGCAAUACAAACACACAAACAGU .......-..(((((.(((.((((((..........)))))).)))...)))))............... ( -3.90, z-score = -0.06, R) >droMoj3.scaffold_6359 778507 68 - 4525533 GGAUUAU-AGGUAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUAGCAAUACAAACACAGGCACAGC .......-..(((((.((((((((((..........))))))))))...)))))............... ( -8.10, z-score = -1.57, R) >droGri2.scaffold_14853 3869580 68 - 10151454 GGAUUAA-AGGUAUUAAAAGUUAUAAAACAACAAGAUUAUGCGCUUAGCAAUACAAACACAGAUACACU .......-..(((((....(((....)))..........(((.....)))...........)))))... ( -6.10, z-score = -0.89, R) >droWil1.scaffold_181096 10865601 68 + 12416693 AGAAGCAAACGUAUUAAAAGUUAUAAAACAACAAGAUUAUGGUUUUAGCAACACACACACACACAGGG- ...................(((.((((((............))))))..)))................- ( -3.80, z-score = 0.03, R) >droPer1.super_53 360698 68 - 525408 GGAUUACGAGGCAUUACGAGCUAUAAAACAACAGGAUUAUGGCUUCGCAAUAGCAAACACACUCACAU- .......(((.......(((((((((..(....)..))))))))).((....)).......)))....- ( -13.90, z-score = -3.98, R) >dp4.chrXL_group1a 7712717 68 + 9151740 GGAUUACGAGGCAUUACGAGCUAUAAAACAACAGGAUUAUGGCUUCGCAAUAGCAAACACACUCACAU- .......(((.......(((((((((..(....)..))))))))).((....)).......)))....- ( -13.90, z-score = -3.98, R) >consensus GGAUUAAGAGGCAUUAAAAGUUAUAAAACAACAAGAUUAUGGCUUUGCCAAAACAAACACACACACA__ .................(((((((((..........)))))))))........................ ( -3.50 = -3.39 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:25 2011