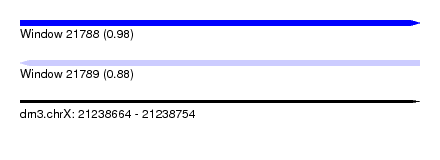
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,238,664 – 21,238,754 |
| Length | 90 |
| Max. P | 0.975271 |

| Location | 21,238,664 – 21,238,754 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.33832 |
| G+C content | 0.44490 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
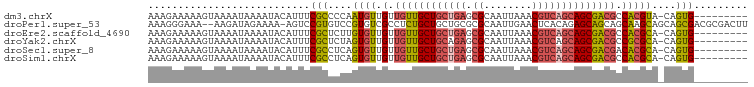
>dm3.chrX 21238664 90 + 22422827 AAAGAAAAAGUAAAAUAAAAUACAUUUCGCCCCAAUGUUGUUGUUGCUGCUGAGCGCAAUUAAACGUCAGCAGCGACGCCACGUA-CAGUG--------- ...((((..(((........))).))))......((((.((.(((((((((((.((........))))))))))))))).)))).-.....--------- ( -24.40, z-score = -2.49, R) >droPer1.super_53 327850 97 + 525408 AAAGGGAAA--AAGAUAGAAAA-AGUCCGUGUCCGUGUCGCCUCUGCUGCUGCGCGCAAUUGAACUCACAGAGCAGCAGCAAGCAGCAGCGACGCGACUU ....(((..--..(((......-.)))....)))(((((((..((((((((((((.....((....))....)).))))).)))))..)))))))..... ( -36.20, z-score = -2.08, R) >droEre2.scaffold_4690 17769249 90 + 18748788 AAAGAAAAAGUAAAAUAAAAUACAUUUCGCUCUUGUGUUGUUGUUGCUGCUGAGCGCAAUUAAACGUCAGCAGCGACGCCACGCA-CAGUG--------- ...((((..(((........))).))))....((((((((.((((((((((((.((........)))))))))))))).)).)))-)))..--------- ( -28.80, z-score = -2.57, R) >droYak2.chrX 20417722 90 + 21770863 AAAGAAAAAGUAAAAUAAAAUACAUUUCGCUCUAGUGUUGUUGUUGCUGCAGAGCGCAAUUAAACGUCAGCAGCGACGCCGCGCA-CAGUG--------- ...((((..(((........))).))))(((...((((.((.((((((((.((.((........)))).)))))))))).)))).-.))).--------- ( -22.10, z-score = -0.09, R) >droSec1.super_8 3546539 90 + 3762037 AAAGAAAAAGUAAAAUAAAAUACAUUUCGCCUCAGUGUUGUUGUUGCUGCUGAGCGCAAUUAAACGUCAGCAGCGACGACACGCA-CAGUG--------- ...((((..(((........))).))))......(((((((((((((((((((.((........))))))))))))))))).)))-)....--------- ( -32.40, z-score = -4.22, R) >droSim1.chrX 16373625 90 + 17042790 AAAGAAAAAGUAAAAUAAAAUACAUUUCGCCUCAGUGUUGUUGUUGCUGCUGAGCGCAAUUAAACGUCAGCAGCGACGCCACGCA-CAGUG--------- ...((((..(((........))).))))......((((((.((((((((((((.((........)))))))))))))).)).)))-)....--------- ( -27.80, z-score = -2.59, R) >consensus AAAGAAAAAGUAAAAUAAAAUACAUUUCGCCCCAGUGUUGUUGUUGCUGCUGAGCGCAAUUAAACGUCAGCAGCGACGCCACGCA_CAGUG_________ ...........................(((....((((.(.((((((((((((.((........)))))))))))))).)))))....)))......... (-20.29 = -20.85 + 0.56)

| Location | 21,238,664 – 21,238,754 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.33832 |
| G+C content | 0.44490 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
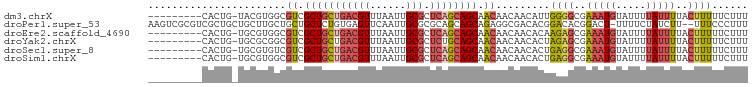
>dm3.chrX 21238664 90 - 22422827 ---------CACUG-UACGUGGCGUCGCUGCUGACGUUUAAUUGCGCUCAGCAGCAACAACAACAUUGGGGCGAAAUGUAUUUUAUUUUACUUUUUCUUU ---------....(-(((((..(((((((((((((((......))).))))))))..(((.....))).))))..))))))................... ( -28.90, z-score = -3.57, R) >droPer1.super_53 327850 97 - 525408 AAGUCGCGUCGCUGCUGCUUGCUGCUGCUCUGUGAGUUCAAUUGCGCGCAGCAGCAGAGGCGACACGGACACGGACU-UUUUCUAUCUU--UUUCCCUUU (((((..((((((.(((((.((((((((....((....))...))).)))))))))).)))))).((....))))))-)..........--......... ( -37.80, z-score = -2.34, R) >droEre2.scaffold_4690 17769249 90 - 18748788 ---------CACUG-UGCGUGGCGUCGCUGCUGACGUUUAAUUGCGCUCAGCAGCAACAACAACACAAGAGCGAAAUGUAUUUUAUUUUACUUUUUCUUU ---------....(-((..((..((.(((((((((((......))).)))))))).))..)).)))(((((..(((((.....)))))..)))))..... ( -28.10, z-score = -2.58, R) >droYak2.chrX 20417722 90 - 21770863 ---------CACUG-UGCGCGGCGUCGCUGCUGACGUUUAAUUGCGCUCUGCAGCAACAACAACACUAGAGCGAAAUGUAUUUUAUUUUACUUUUUCUUU ---------....(-((...(..((.(((((.(((((......))).)).))))).))..)..))).((((..(((((.....)))))..))))...... ( -20.00, z-score = 0.24, R) >droSec1.super_8 3546539 90 - 3762037 ---------CACUG-UGCGUGUCGUCGCUGCUGACGUUUAAUUGCGCUCAGCAGCAACAACAACACUGAGGCGAAAUGUAUUUUAUUUUACUUUUUCUUU ---------....(-(((((.((((((((((((((((......))).)))))))).....((....)).))))).))))))................... ( -28.80, z-score = -3.06, R) >droSim1.chrX 16373625 90 - 17042790 ---------CACUG-UGCGUGGCGUCGCUGCUGACGUUUAAUUGCGCUCAGCAGCAACAACAACACUGAGGCGAAAUGUAUUUUAUUUUACUUUUUCUUU ---------....(-(((((..(((((((((((((((......))).)))))))).....((....)).))))..))))))................... ( -28.40, z-score = -2.62, R) >consensus _________CACUG_UGCGUGGCGUCGCUGCUGACGUUUAAUUGCGCUCAGCAGCAACAACAACACUGAGGCGAAAUGUAUUUUAUUUUACUUUUUCUUU .......................((.(((((((((((......))).)))))))).)).........((((..(((((.....)))))..))))...... (-16.44 = -17.25 + 0.81)
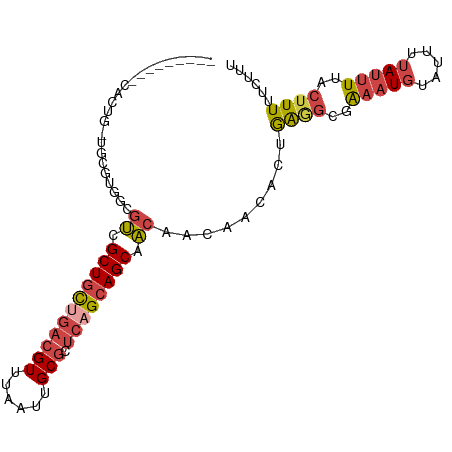
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:20 2011