
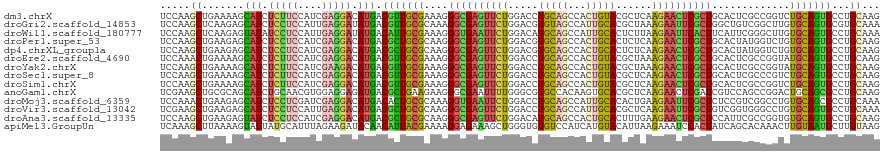
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,225,592 – 21,225,712 |
| Length | 120 |
| Max. P | 0.754300 |

| Location | 21,225,592 – 21,225,712 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Shannon entropy | 0.48905 |
| G+C content | 0.53750 |
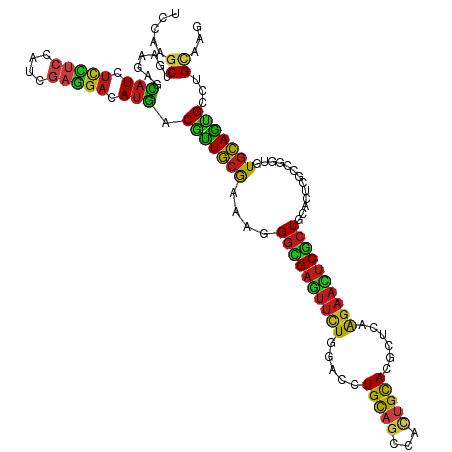
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -25.36 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.66 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21225592 120 + 22422827 UCCAAGCUGAAAAGCAUCUCUUCCAUCGAGGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGUACGCUCAAGAACUCGCUGCACUCGCCGGUCUGCAGUGCCUGCAAG .....(((....)))...(((((....))))).......(((((....(((((((((((..(.(((((...))))).)..).))))))))))(((((.((......))))))).))))). ( -40.70, z-score = -0.76, R) >droGri2.scaffold_14853 7172892 120 + 10151454 UCCAAGCUCAAGAGCAUCUCCUCCAUUGAGGAUAUGACGUUGCGCAAAGGCGAGUUCUGGACGUGUAGCCAUUGCACGCUAAAGAAUUCGCUGGCUGUCGGCUUGUGCAGUGCGUGCAAA .....((.......(((.(((((....))))).)))(((((((((((.((((((((((((.(((((((...)))))))))..)))))))))).((.....)))))))))..))))))... ( -43.70, z-score = -1.32, R) >droWil1.scaffold_180777 2091336 120 + 4753960 UCCAAGCUCAAGAGUAUAUCCUCCAUUGAGGAUAUGACAUUGCGAAAGGGUGAAUUCUGGACAUGCAGCCAUUGCACUCUUAAGAAUUCACUUCAUUCGGGCUUGUGCAGUGCCUGCAAA ..(((((((((...(((((((((....)))))))))...)))((((.((((((((((((((..(((((...))))).)))..)))))))))))..))))))))))(((((...))))).. ( -42.90, z-score = -3.30, R) >droPer1.super_53 307528 120 + 525408 UCCAAGCUGAAGAGCAUCUCCUCCAUCGAGGACAUGACGCUGCGCAAGGGCGAGUUCUGGACGUGCAGCCACUGCACUCUCAAGAACUCGCUGCACUAUGGUCUGUGCAGUGCCUGCAAG .....((...((..(((.(((((....))))).)))...))..))..((((((((((((((.((((((...)))))))))..)))))))(((((((........)))))))))))..... ( -49.40, z-score = -2.16, R) >dp4.chrXL_group1a 7660585 120 - 9151740 UCCAAGCUGAAGAGCAUCUCCUCCAUCGAGGACAUGACGCUGCGCAAGGGCGAGUUCUGGACGUGCAGCCACUGCACUCUCAAGAACUCGCUGCACUAUGGUCUGUGCAGUGCCUGCAAG .....((...((..(((.(((((....))))).)))...))..))..((((((((((((((.((((((...)))))))))..)))))))(((((((........)))))))))))..... ( -49.40, z-score = -2.16, R) >droEre2.scaffold_4690 17756620 120 + 18748788 UCCAAACUGAAAAGCAUCUCUUCCAUCGAGGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGUACGCUAAAGAACUCGCUGCACUCGCCGGUAUGCAGUGCCUGCAAG .....((((....((((.(((((....))))).))).)...((((...((((((((((((.(.(((((...))))).)))..))))))))))....)))))))).(((((...))))).. ( -37.10, z-score = -0.72, R) >droYak2.chrX 20403597 120 + 21770863 UCCAAGCUGAAAAGCAUCUCUUCCAUCGAAGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGUACGCUAAAGAACUCGCUGCACUCGCCGGUAUGCAGUGCCUGCAAG .....(((....)))...(((((....))))).......(((((....((((((((((((.(.(((((...))))).)))..))))))))))(((((.((......))))))).))))). ( -39.10, z-score = -1.15, R) >droSec1.super_8 3533351 120 + 3762037 UCCAAGCUGAAAAGCAUCUCUUCCAUCGAGGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGUACGCUCAAGAACUCGCUGCACUCGCCCGUCUGCAGUGCCUGCAAG .....(((....)))...(((((....)))))...((((..((((...(((((((((((..(.(((((...))))).)..).))))))))))....)))).))))(((((...))))).. ( -42.80, z-score = -1.70, R) >droSim1.chrX 16359856 120 + 17042790 UCCAAGCUGAAAAGCAUCUCUUCCAUCGAGGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGUACGCUCAAGAACUCGCUGCACUCGCCGGUCUGCAGUGCCUGCAAG .....(((....)))...(((((....))))).......(((((....(((((((((((..(.(((((...))))).)..).))))))))))(((((.((......))))))).))))). ( -40.70, z-score = -0.76, R) >anoGam1.chrX 15359139 120 + 22145176 UCGAAGCUGCGCAGCAUCUGCAACGUGGAGGAGAUGACGCUGAAGAAGGGCGAAUUUUGGGCGUGCCACAAGUGCACGCUCAAGAACUCGAUCGUCCAGCCGGACUGCAGCGCCUGCAAG .....((.((((.(((((((...(.((((.((..(((((((.......))))..(((((((((((((....).))))))))))))..))).)).))))).)))).))).))))..))... ( -46.30, z-score = -0.97, R) >droMoj3.scaffold_6359 2407344 120 - 4525533 UCCAAACUGAAGAGCAUCUCCUCGAUCGAGGACAUGACACUGCGCAAAGGUGAAUUCUGGACCUGCAGCCAUUGCACACUGAAGAAUUCGCUCUCCGUCGGCCUGUGCAGCGCCUGCAAA ((((..........(((.(((((....))))).))).((((.......)))).....))))..(((((.(.((((((((((((((......)))...))))..))))))).).))))).. ( -35.00, z-score = -0.21, R) >droVir3.scaffold_13042 4308020 120 + 5191987 UCGAAGCUGAAGAGCAUCUCCUCCAUUGAGGACAUGACGCUGCGCAAGGGCGAGUUCUGGACCUGCAGCCAUUGCACGCUCAAGAAUUCGCUGUCGGUGGGCCUGUGCAGUGCCUGCAAA .....((.......(((.(((((....))))).))).(((((((((..(((((((((((..(.(((((...))))).)..).))))))))))...((....)))))))))))...))... ( -44.70, z-score = -0.34, R) >droAna3.scaffold_13335 2140480 120 + 3335858 UCCAAGCUGAAGAGUAUCUCCUCCAUCGAGGACAUGACGCUGCGCAAGGGCGAGUUCUGGACAUGCAGCCACUGCACUUUGAAGAACUCGCUCCAUUCGCCGGUGUGCAGUGCCUGCAAG .....((.......(((.(((((....))))).))).(((((((((.(((((((((((.((..(((((...)))))..))..)))))))))))..........)))))))))...))... ( -43.80, z-score = -1.28, R) >apiMel3.GroupUn 6259056 120 + 399230636 UCAAAGCUUAAAAGUAUUAUGCAUUUAGAAGAUACAACAUUACGAAAAGGAGAAAGCUGGGUGUGUCCAUCAUGUACAUUAAGAAAUCCACUAUCAGCACAAACUUGUAAUGCUUGUAAG .....((.(((.....))).))..........((((((((((((...((......(((((..(((....((.((.....)).))....)))..))))).....))))))))).))))).. ( -17.90, z-score = 0.89, R) >consensus UCCAAGCUGAAGAGCAUCUCCUCCAUCGAGGACAUGACGUUGCGAAAGGGCGAGUUCUGGACCUGCAGCCACUGCACGCUCAAGAACUCGCUGCACUCGCCGGUGUGCAGUGCCUGCAAG .....((.......(((.(((((....))))).))).(((((((....((((((((((.....(((((...)))))......)))))))))).............)))))))...))... (-25.36 = -24.52 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:19 2011