
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,174,229 – 21,174,463 |
| Length | 234 |
| Max. P | 0.646639 |

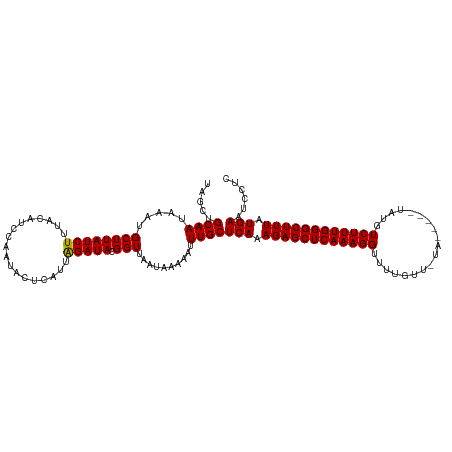
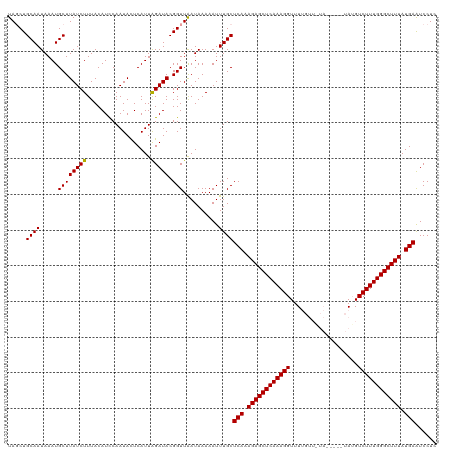
| Location | 21,174,229 – 21,174,344 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.01 |
| Shannon entropy | 0.15095 |
| G+C content | 0.31793 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.04 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21174229 115 - 22422827 UAGCUGCAAUAAAUGCUUAUCUUUACAUUCAAUAUUCAUUGGAUACGGUUAAUAAAAAUUUGCUCAAAGAGCUCAAAGGUUUUGUUUUA-----UAAGUCUUUGGGCUUUAUGAGUCCUC ((((((((.....)))..........(((((((....)))))))..)))))..........(((((.(((((((((((..((.......-----.))..))))))))))).))))).... ( -29.70, z-score = -3.40, R) >droSec1.super_8 3481563 120 - 3762037 UAUCUGCAAUAAAUGCUUAUCUUUACAUCCAAUACUCAUUAGAUACGGUUGAUAAAAAUUUGCUCAAAGAGCUCAAAGGUUUUGUUGUAUAUAGUAUGUCUUUGGGCUUUAUGAAUCCUC .....((((...((..(((((..(..(((.(((....))).)))...)..)))))..))))))(((.((((((((((((..((((.....))))....)))))))))))).)))...... ( -21.20, z-score = -0.31, R) >droSim1.chrX_random 5481208 108 - 5698898 UAGCUGCAAUAAAUGCUUAUCUUUACAUCCAAUACUCAUUAGAUACGGUUAAUAAAAAUUUGCUCAAAGAGCUCAAAGGUUU------------UAUGUCUUUGGGCUUUAUGAAUCCUC ((((((((.....))).(((((..................))))).)))))............(((.((((((((((((...------------....)))))))))))).)))...... ( -21.07, z-score = -1.37, R) >consensus UAGCUGCAAUAAAUGCUUAUCUUUACAUCCAAUACUCAUUAGAUACGGUUAAUAAAAAUUUGCUCAAAGAGCUCAAAGGUUUUGUU_UA_____UAUGUCUUUGGGCUUUAUGAAUCCUC .....((((.....((((((((..................))))).)))..........))))(((.((((((((((((...................)))))))))))).)))...... (-19.26 = -19.04 + -0.22)

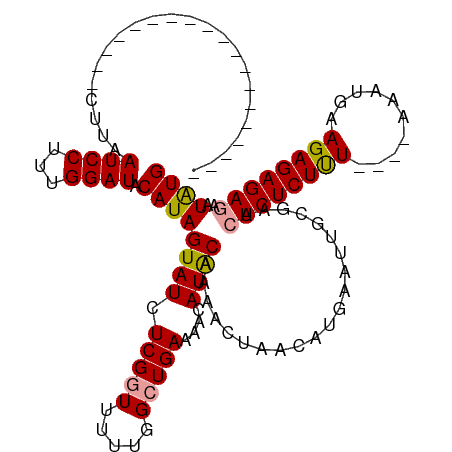
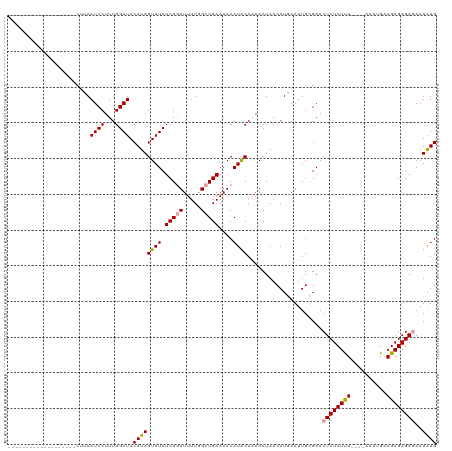
| Location | 21,174,344 – 21,174,463 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Shannon entropy | 0.26784 |
| G+C content | 0.33592 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21174344 119 - 22422827 UAUAACCACCAAAAAAUUACCGAAUCCUUUGGAUACAUAGUAUCUCGUUUUUU-GCUGAAAACAUGCAUAACUAACAUCAAUUGGGAACUCUCUCUCUUGAAAUGAAGAGAGAGAAUGUG ......(((..........((((.......((((((...)))))).((((((.-...))))))..................))))....((((((((((......))))))))))..))) ( -25.80, z-score = -2.60, R) >droSec1.super_8 3481683 97 - 3762037 -------------------CUUAAUCCUUUGGAUACAUAGUAUCUCGGUUUUUGGCUGAAAACAUACAAAACUAACAUGAAUUGCGAACUCUCUUU----AAAUGAAGAGAGACAAUAUG -------------------....((((...)))).(((((((..((((((((((..((....))..))))...))).)))..)))...((((((((----....))))))))....)))) ( -18.00, z-score = -1.84, R) >droSim1.chrX_random 5481316 96 - 5698898 -------------------CUUAAUCCUUUGGAUACAUAGUAUCUCGGUUUUUGGCUGAAAACAUACAAAACUAACAUGAAUUGCGAACUCUCUUU----AAAUG-AGAGAGAGAAUAUG -------------------....((((...)))).(((((((..((((((((((..((....))..))))...))).)))..)))...((((((((----.....-))))))))..)))) ( -17.80, z-score = -1.34, R) >consensus ___________________CUUAAUCCUUUGGAUACAUAGUAUCUCGGUUUUUGGCUGAAAACAUACAAAACUAACAUGAAUUGCGAACUCUCUUU____AAAUGAAGAGAGAGAAUAUG .......................((((...)))).((((((((.(((((.....)))))....)))).....................((((((((..........))))))))..)))) (-13.69 = -13.70 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:14 2011