
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,172,723 – 21,172,821 |
| Length | 98 |
| Max. P | 0.947431 |

| Location | 21,172,723 – 21,172,816 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.26994 |
| G+C content | 0.38073 |
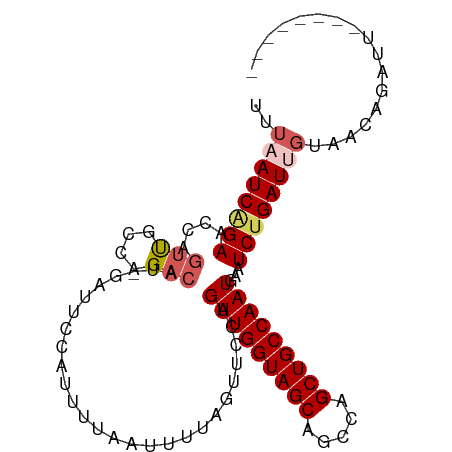
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -18.18 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
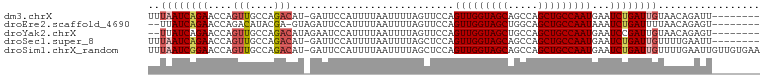
>dm3.chrX 21172723 93 - 22422827 UUUAAUCAGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUAACAGAUU-------- ..((((((((....(((....)))..-........................(((((((((.....)))))))))...)))))))).........-------- ( -22.00, z-score = -2.54, R) >droEre2.scaffold_4690 17703134 91 - 18748788 --UUAUCAGAACCAGACAUACGA-GUAGAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCUGGCAGCUGCCAAUAAAUCUGAUUUUAACAGAGU-------- --..((((((....(((.((.((-.((((......)))).)).)))))...(((((((((.....)))))))))...))))))...........-------- ( -19.00, z-score = -1.71, R) >droYak2.chrX 20349487 92 - 21770863 --UUAUCAGAACCAGUUGCCAGACAUAGAAUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCUGCCAGCUGCCAAUGAAUCCGAUUGUAACAGAGU-------- --............(((((..(((.((((((........)))))))))...(((((((((.....)))))))))..........))))).....-------- ( -19.50, z-score = -1.95, R) >droSec1.super_8 3480001 93 - 3762037 UUUAAUCAGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGCUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUUUUGAAUU-------- ..((((((((....(((....)))..-........................(((((((((.....)))))))))...)))))))).........-------- ( -22.00, z-score = -2.02, R) >droSim1.chrX_random 5501430 101 - 5698898 UUUAAUCGGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGCUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUUUUGAAUUGUUGUGAA .....(((.(((.((((.(.(((((.-(((((...................(((((((((.....))))))))))))))....))))).)))))))).))). ( -23.50, z-score = -1.12, R) >consensus UUUAAUCAGAACCAGUUGCCAGACAU_GAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUAACAGAUU________ ..((((((((....(((....)))...........................(((((((((.....)))))))))...))))))))................. (-18.18 = -19.10 + 0.92)
| Location | 21,172,723 – 21,172,821 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.27019 |
| G+C content | 0.37756 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -18.22 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
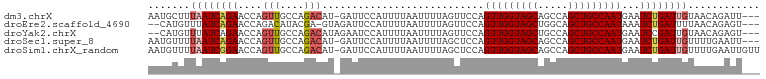
>dm3.chrX 21172723 98 - 22422827 AAUGCUUUAAUCAGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUAACAGAUU--- ..((...((((((((....(((....)))..-........................(((((((((.....)))))))))...))))))))...))....--- ( -22.40, z-score = -1.83, R) >droEre2.scaffold_4690 17703134 96 - 18748788 --CAUGUUUAUCAGAACCAGACAUACGA-GUAGAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCUGGCAGCUGCCAAUAAAUCUGAUUUUAACAGAGU--- --..((((.((((((....(((.((.((-.((((......)))).)).)))))...(((((((((.....)))))))))...))))))...))))....--- ( -22.30, z-score = -2.31, R) >droYak2.chrX 20349487 97 - 21770863 --CAUGUUUAUCAGAACCAGUUGCCAGACAUAGAAUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCUGCCAGCUGCCAAUGAAUCCGAUUGUAACAGAGU--- --..((((.(((.((((..(((....))).((((((........))))))))))..(((((((((.....)))))))))......)))...))))....--- ( -21.20, z-score = -1.96, R) >droSec1.super_8 3480001 98 - 3762037 AAUGUUUUAAUCAGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGCUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUUUUGAAUU--- .......((((((((....(((....)))..-........................(((((((((.....)))))))))...)))))))).........--- ( -22.00, z-score = -1.60, R) >droSim1.chrX_random 5501435 101 - 5698898 AAUGUUUUAAUCGGAACCAGUUGCCAGACAU-GAUUCCAUUUUAAUUUUAGCUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUUUUGAAUUGUU .......(((((((((.(((((....))).)-).))))..................(((((((((.....)))))))))......)))))............ ( -22.50, z-score = -1.06, R) >consensus AAUGUUUUAAUCAGAACCAGUUGCCAGACAU_GAUUCCAUUUUAAUUUUAGUUCCAGUUGGUAGCAGCCAGCUGCCAAUGAAUCUGAUUGUAACAGAUU___ .......((((((((....(((....)))...........................(((((((((.....)))))))))...))))))))............ (-18.22 = -19.14 + 0.92)
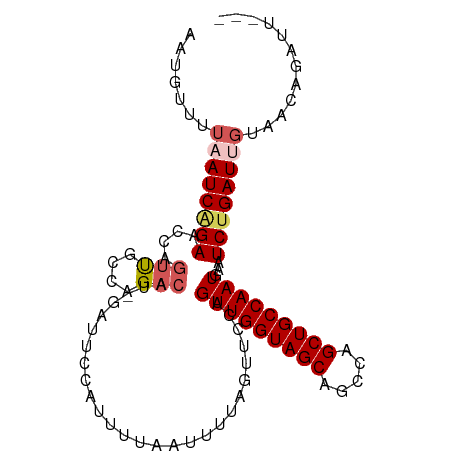
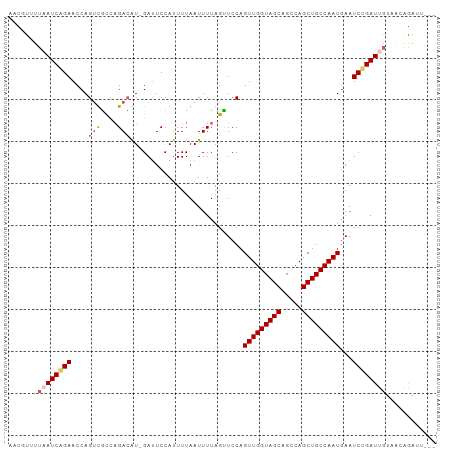
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:12 2011