| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,263,069 – 12,263,172 |
| Length | 103 |
| Max. P | 0.999769 |

| Location | 12,263,069 – 12,263,172 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.36 |
| Shannon entropy | 0.62892 |
| G+C content | 0.42115 |
| Mean single sequence MFE | -19.16 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
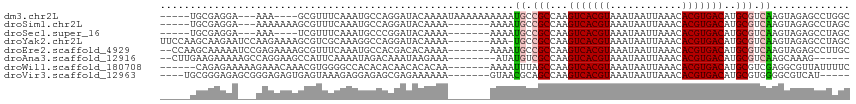
>dm3.chr2L 12263069 103 + 23011544 -----UGCGAGGA---AAA----GCGUUUCAAAUGCCAGGAUACAAAAUAAAAAAAAAAUGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUGGC -----.((.....---...----)).........((((((.(((....(......)...((.(((...(((((((............)))))))..))).)).)))...)))))) ( -24.10, z-score = -2.16, R) >droSim1.chr2L 12064435 100 + 22036055 -----UGCGAGGA---AAAAAAAGCGUUUCAAAUGCCAGGAUACAAAA-------AAAAUGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUAGC -----.((.(((.---.......((((.....))))............-------....((.(((...(((((((............)))))))..))).)).......))).)) ( -19.20, z-score = -0.91, R) >droSec1.super_16 453261 96 + 1878335 -----UGCGAGGA---AAA----UCGUUUCAAAUGCCCGGAUACAAAA-------AAAAUGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUAGC -----.((.(((.---...----((((.((.........)).))....-------....((.(((...(((((((............)))))))..))).))....)).))).)) ( -18.60, z-score = -0.69, R) >droYak2.chr2L 8692578 107 + 22324452 UUCCAAGCAAGAAUCCAAGAAAAGCGUCGCAAAGGCCAGGAUACAAAA-------AAA-UGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUAGC ......((.((..((...(.....)(.((((..(((..((((......-------..)-).)))))..(((((((............))))))).)))).).....))..)).)) ( -19.70, z-score = -1.24, R) >droEre2.scaffold_4929 13486541 106 - 26641161 --CCAAGCAAAAAUCCGAGAAAAGCGUUUCAAAUGCCACGACACAAAA-------AAAAUGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUUGC --....((((...((........((((.....))))............-------....((.(((...(((((((............)))))))..))).))....))...)))) ( -17.90, z-score = -0.80, R) >droAna3.scaffold_12916 3052214 99 - 16180835 --CUUGAAGAAAAAGCCAGGAAGCCAUUCAAAAUAGACAAAUAAGAAA--------AUAUGUCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGCAAAG------ --(((((.(((...((......))..)))......((((.((......--------)).))))((...(((((((............)))))))..)).))))).....------ ( -17.90, z-score = -2.70, R) >droWil1.scaffold_180708 2611221 102 - 12563649 ------CAGAGAAAAAGAAACAAACGUGGGGCCACACACAACACACAA-------AAAAUUUAGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCGAGGCGUUAUUUUC ------..........((((..((((((....))).............-------....((..((((.(((((((............))))))).)).))..))..)))..)))) ( -17.20, z-score = -0.62, R) >droVir3.scaffold_12963 2868054 99 - 20206255 ----UGCGGGAGAGCGGGAGAGUGAGUAAAGAGGAGAGCGAGAAAAAA-------GUAACGCAGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUGGGGCGUCAU----- ----.((......)).................................-------...((((.((((.(((((((............))))))).)).))...))))...----- ( -18.70, z-score = -0.21, R) >consensus _____UGCGAGAA___AAAAAAAGCGUUUCAAAUGCCAGGAUACAAAA_______AAAAUGCCGCCAAGUCACGUAAAUAAUUAAACACGUGACAUGCGUCAAGUAGAGCCUAGC ...........................................................((.(((...(((((((............)))))))..))).))............. (-11.50 = -11.70 + 0.20)
| Location | 12,263,069 – 12,263,172 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.36 |
| Shannon entropy | 0.62892 |
| G+C content | 0.42115 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -14.75 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
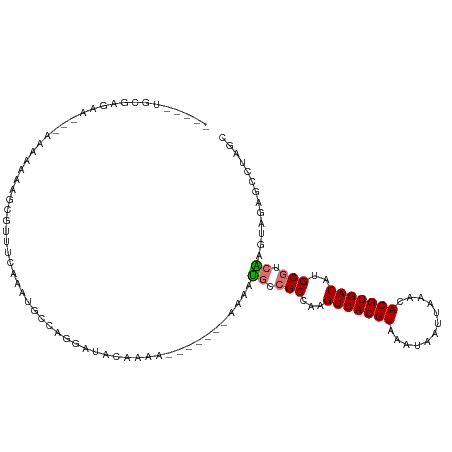
>dm3.chr2L 12263069 103 - 23011544 GCCAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCAUUUUUUUUUUAUUUUGUAUCCUGGCAUUUGAAACGC----UUU---UCCUCGCA----- ((((((..........(((..((((((((..........))))))))...)))(((..............)))..)))))).........((----...---.....)).----- ( -28.64, z-score = -3.55, R) >droSim1.chr2L 12064435 100 - 22036055 GCUAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCAUUUU-------UUUUGUAUCCUGGCAUUUGAAACGCUUUUUUU---UCCUCGCA----- ((((((..........(((..((((((((..........))))))))...)))(((....-------...)))..)))))).........((.......---.....)).----- ( -26.20, z-score = -2.86, R) >droSec1.super_16 453261 96 - 1878335 GCUAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCAUUUU-------UUUUGUAUCCGGGCAUUUGAAACGA----UUU---UCCUCGCA----- ((.((((((.......(((..((((((((..........))))))))...)))(((....-------...)))....))))....((((...----.))---)))).)).----- ( -24.90, z-score = -1.90, R) >droYak2.chr2L 8692578 107 - 22324452 GCUAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCA-UUU-------UUUUGUAUCCUGGCCUUUGCGACGCUUUUCUUGGAUUCUUGCUUGGAA ((((((..........(((..((((((((..........))))))))...)))(((-...-------...)))..))))))((..((((..((......))....))))..)).. ( -29.90, z-score = -2.74, R) >droEre2.scaffold_4929 13486541 106 + 26641161 GCAAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCAUUUU-------UUUUGUGUCGUGGCAUUUGAAACGCUUUUCUCGGAUUUUUGCUUGG-- ((((((.(((......(((..((((((((..........))))))))...)))((.((((-------....((((....))))..)))).)).......))).))))))....-- ( -29.30, z-score = -2.27, R) >droAna3.scaffold_12916 3052214 99 + 16180835 ------CUUUGCUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGACAUAU--------UUUCUUAUUUGUCUAUUUUGAAUGGCUUCCUGGCUUUUUCUUCAAG-- ------.....(((((.((..((((((((..........))))))))...))((((.((--------......)).))))......(((.((((....))))..))).)))))-- ( -24.70, z-score = -4.12, R) >droWil1.scaffold_180708 2611221 102 + 12563649 GAAAAUAACGCCUCGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCUAAAUUUU-------UUGUGUGUUGUGUGUGGCCCCACGUUUGUUUCUUUUUCUCUG------ (((((.(((((..((((((((((((((((..........)))))))).............-------..)))))))).))(((....)))....)))...)))))....------ ( -24.46, z-score = -2.36, R) >droVir3.scaffold_12963 2868054 99 + 20206255 -----AUGACGCCCCACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCUGCGUUAC-------UUUUUUCUCGCUCUCCUCUUUACUCACUCUCCCGCUCUCCCGCA---- -----.((((((.(((.....((((((((..........)))))))).)))..)))))).-------.................................((......)).---- ( -20.30, z-score = -3.60, R) >consensus GCUAGGCUCUACUUGACGCAUGUCACGUGUUUAAUUAUUUACGUGACUUGGCGGCAUUUU_______UUUUGUAUCCUGGCAUUUGAAACGCUUUUUUU___UCCUCGCA_____ ................(((..((((((((..........))))))))...))).............................................................. (-14.75 = -15.00 + 0.25)

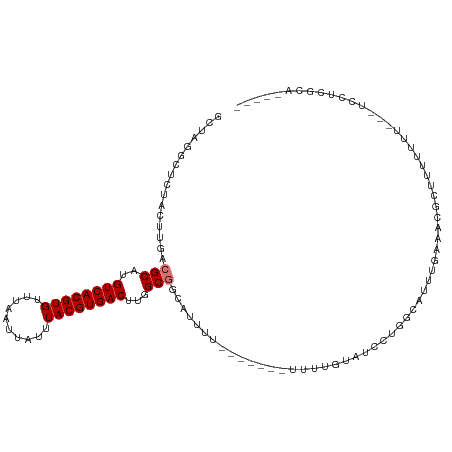
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:41 2011