| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,117,255 – 21,117,358 |
| Length | 103 |
| Max. P | 0.964379 |

| Location | 21,117,255 – 21,117,358 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.16 |
| Shannon entropy | 0.76077 |
| G+C content | 0.52947 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -10.66 |
| Energy contribution | -10.79 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
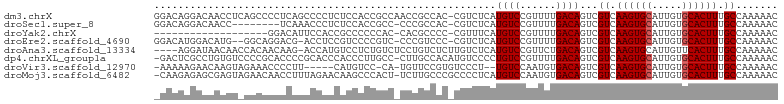
>dm3.chrX 21117255 103 - 22422827 GGACAGGACAACCUCAGCCCCUCAGCCCCUCUCCACCGCCAACCGCCAC-CGUCUCAUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC (((((((((.......((......))...........((.....))...-.))))..)))))((((((((....)))((((((.....)))))).....))))) ( -21.90, z-score = -0.80, R) >droSec1.super_8 3423610 94 - 3762037 GGACAGGACAACC--------UCAAACCCUCUCCACCGCC-CCCGCCAC-CGUCUCAUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC ((((((((.....--------..........)))...((.-...))...-.......)))))((((((((....)))((((((.....)))))).....))))) ( -18.46, z-score = -0.39, R) >droYak2.chrX 20291120 83 - 21770863 -------------------GGACAUUCCACCGCCCCCCAC-CACGCCCC-CGUUUCAUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC -------------------((((((...((.((.......-...))...-.))...))))))((((((((....)))((((((.....)))))).....))))) ( -16.20, z-score = -1.06, R) >droEre2.scaffold_4690 17647210 99 - 18748788 GGACAUGGACAUG--GGCAGGACG-ACCUCCGUCCCCGUC-CCCGUCCC-CGUCUCAUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC ((((((((((..(--(((.(((((-...........))))-)..)))).-.))).)))))))((((((((....)))((((((.....)))))).....))))) ( -38.20, z-score = -3.42, R) >droAna3.scaffold_13334 444251 99 + 1562580 ----AGGAUAACAACCACAACAAG-ACCAUGUCCUCUGUCUCCUGUCUCUUGUCUCAUGUCCGUUCUGACAGUCGUCAAGUGCAUUGUUCACUUUGCCAAAAAC ----.(((((((((..(((...((-((..........))))..)))...))))....)))))....((((....)))).(.(((..(....)..))))...... ( -14.60, z-score = 0.89, R) >dp4.chrXL_group1a 33633 102 + 9151740 -GACUCGCCUGUGUCCCCGCACCCCGCACCCACCCUUGCC-CUUGCCACAUGUCCCCUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC -(((..((.((((.....(((....(((........))).-..))))))).))...(((((......)))))..)))((((((.....)))))).......... ( -21.00, z-score = -1.40, R) >droVir3.scaffold_12970 4035363 94 + 11907090 -AAAAAGAACAAGUAGAAACCCCUU-----CAUGUCC-CA-UGUUCCGUGUCCCU--UGUCCAAUGUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC -.....(.(((((..((.((.....-----((((...-))-))....)).)).))--))).)....((((....))))(((((.....)))))........... ( -14.90, z-score = -0.67, R) >droMoj3.scaffold_6482 1667969 102 - 2735782 -CAAGAGAGCGAGUAGAACAACCUUUAGAACAAGCCCACU-UCUUGCCCGCCCCUCAUGUCCAAUGUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC -...(((.(((.((((((....(((......))).....)-)).))).)))..))).((((......))))...((.((((((.....)))))).))....... ( -20.90, z-score = -0.79, R) >consensus _GACAGGACAACGU_GACACCACAUACCACCACCCCCGCC_CCCGCCAC_CGUCUCAUGUCCGUUUUGACAGUCGUCAAGUGCAUUGUGCACUUUGCCAAAAAC .........................................................((((......))))...((.((((((.....)))))).))....... (-10.66 = -10.79 + 0.12)

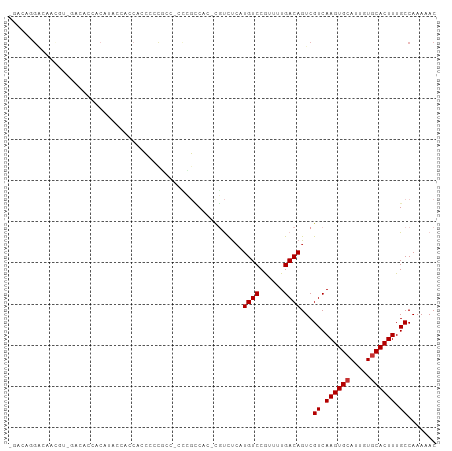
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:05 2011