| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,108,802 – 21,108,951 |
| Length | 149 |
| Max. P | 0.956652 |

| Location | 21,108,802 – 21,108,951 |
|---|---|
| Length | 149 |
| Sequences | 10 |
| Columns | 156 |
| Reading direction | reverse |
| Mean pairwise identity | 72.44 |
| Shannon entropy | 0.57988 |
| G+C content | 0.54363 |
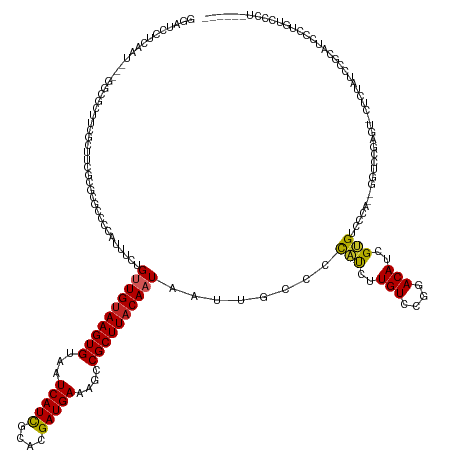
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
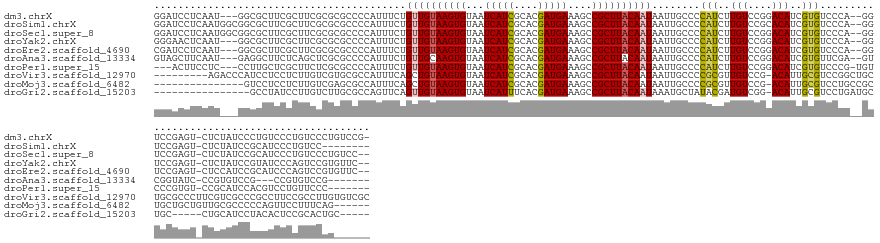
>dm3.chrX 21108802 149 - 22422827 GGAUCCUCAAU---GGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCA--GGUCCGAGU-CUCUAUCCCUGUCCCUGUCCCUGUCCG- ((((.....((---((.((..(((.....))).........(((((((((((...(((((....)))))....)))))))))))...)).))))...))))(((((..(.(...((--((..((.(.-.......).))..)))).)).))))).- ( -40.40, z-score = -2.72, R) >droSim1.chrX 16278637 145 - 17042790 GGAUCCUCAAUGGCGGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGCACAUCGUGUCCCA--GGUCCGAGU-CUCUAUCCGCAUCCCUGUCC-------- (((..((((((((.(((((..........))))))))))..(((((((((((...(((((....)))))....)))))))))))........((((.(..(((.....)))..).)--)))..))).-.)))................-------- ( -41.60, z-score = -2.40, R) >droSec1.super_8 3415319 151 - 3762037 GGAUCCUCAAUGGCGGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCA--GGUCCGAGU-CUCUAUCCGCAUCCCUGUCCCUGUCC-- ((((....(((((.(((((..........))))))))))..(((((((((((...(((((....)))))....))))))))))).............))))((((.....))))((--((..((...-...............))..))))...-- ( -44.77, z-score = -2.63, R) >droYak2.chrX 20281786 148 - 21770863 GGGAACUCAAU---GGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCA--GGUCCGAGU-CUCUAUCCGUAUCCCAGUCCGUGUUC-- ((((((.((((---(((((..........))))).......(((((((((((...(((((....)))))....)))))))))))))))......((((.((((((.....))))..--))..)))).-........)).))))...........-- ( -41.20, z-score = -1.79, R) >droEre2.scaffold_4690 17638947 148 - 18748788 CGAUCCUCAAU---GGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCA--GGUCCGAGU-CUCCAUCCGCAUCCCAGUCCGUGUUC-- .(((...((((---(((((..........))))).......(((((((((((...(((((....)))))....)))))))))))))))....))).....(((((.(((.(.(...--.).))))((-........))......))))).....-- ( -37.30, z-score = -1.44, R) >droAna3.scaffold_13334 435164 140 + 1562580 GUAGCUUCAAU---GAGGCUUCUCAGCUCGCGCCCCAUUUCUGUUGCAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUUCGA--GUCGGUAUC-CCGUGUCCG---CCGUGUCCG------- (((((...(((---(.(((............))).))))...)))))(((((...(((((....)))))....)))))......................(((((((.(((..((.--..(((....-)))))..))---).)))))))------- ( -36.30, z-score = -0.90, R) >droPer1.super_15 662506 141 + 2181545 ---ACUUCCUC---CCUUGCUCGCUUCUCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCG-UGUCCCGUGU-CCGCAUCCACGUCCUGUUCCC------- ---........---...(((.(((....((((.........(((((((((((...(((((....)))))....))))))))))).................((((.....))))))-))....))).-..)))................------- ( -30.90, z-score = -2.91, R) >droVir3.scaffold_12970 7853374 146 - 11907090 ---------AGACCCAUCCUCCUCUUGUCGUGCGCCAUUUCAGCUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCGCGUUGUCCG-ACAUUGCGUCCGGCUGCUGCGCCCUUCGUCGCCCGCCUUCCGCCUUGUGUCGC ---------.(((.((.........(((((.((((.....(((.((((((((...(((((....)))))....))))))))....)))....))))....))-)))..(((...(((.((.(((.....))).))..)))...)))..)).))).. ( -38.00, z-score = -0.94, R) >droMoj3.scaffold_6482 1047423 134 + 2735782 ---------------GUCCUCCUCUUGUCGAGCGCCAUUUCAGCUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCGCGUUGUCCG-ACAUUGCGUCCUGCCGCUGCUGCUGUUGCGCCCCCAGUUCCUUUCAG------ ---------------.........(((..(.((((.((..((((((((((((...(((((....)))))....))))))))...........(((..((..(-((.....)))..))))).))))..)).)))))..)))..........------ ( -34.20, z-score = -1.75, R) >droGri2.scaffold_15203 807841 129 + 11997470 ----------------GCCUAUCCUUGUCUUGCGCCAGUUCAGUUGUAAGUGUAAUCAUUUCACGAUGAAAGCCGCUUACAAUAAAUGCUAUACGAUGUCGG-ACAUUGCGUCCUGAUGCUGC-----CUGCAUCCUACACUCCGCACUGC----- ----------------..........((..((((..(((...((((((((((...(((((....)))))....))))))))))....)))....(((((.((-((.....)))).(((((...-----..)))))..))).))))))..))----- ( -30.60, z-score = -0.67, R) >consensus GGAUCCUCAAU___GGCGCUUCGCUUCGCGCGCCCCAUUUCUGUUGUAAGUGUAAUCAUCGCACGAUGAAAGCCGCUUACAAUAAUUGCCCCAUCUUGUCCGGACAUCGUGUCCCA__GGUCCGAGU_CUCUAUCCGCAUCCCUGUCCCU______ ..........................................((((((((((...(((((....)))))....))))))))))........(((..(((....)))..)))............................................. (-17.57 = -17.30 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:04 2011