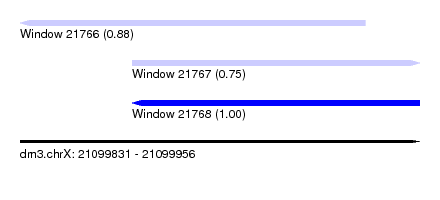
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,099,831 – 21,099,956 |
| Length | 125 |
| Max. P | 0.999009 |

| Location | 21,099,831 – 21,099,939 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Shannon entropy | 0.42746 |
| G+C content | 0.38207 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.56 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
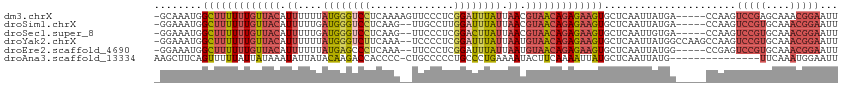
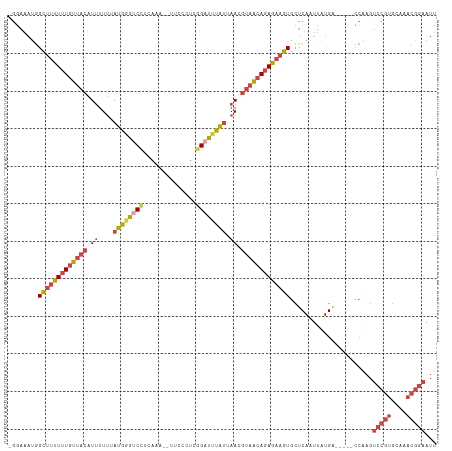
>dm3.chrX 21099831 108 - 22422827 -GCAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCAAAAGUUCCCUCGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGA-----CCAAGUCCGAGCAAACGGAAUU -.......(((((((((((((.((....((((((((..............)))))))).)).)))))))))))))............-----.....((((......))))... ( -24.14, z-score = -1.16, R) >droSim1.chrX 16269732 106 - 17042790 -GGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCAAG--UUGCCUUGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGA-----CCAAGUCCGUGCAAACGGAAUU -((.....(((((((((((((....(((((((((((.....--.......))))))))))).)))))))))))))..(((....)))-----))...(((((....)))))... ( -31.20, z-score = -2.74, R) >droSec1.super_8 3406493 106 - 3762037 -GGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCAAG--UUCCCUCGGACUUAUUAACGUAACAGAGAAGUGCUCAAUUGUGA-----CCAAGUCCGUGCAAACGGAAUU -((.....(((((((((((((....(((((((((((.....--.......))))))))))).)))))))))))))((......))..-----))...(((((....)))))... ( -33.20, z-score = -2.89, R) >droYak2.chrX 20272030 111 - 21770863 -GGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCUUCAAA--UCCCCUCGGAUUUAUUAAUGUAACAGAGAAGUGCUCAAUUAUGGCCAAGCCAAGUCCGUGCAAACGGAAUU -.......((((((((((((((((....((((((((.....--.......)))))))).)))))))))))))))).........((((...))))..(((((....)))))... ( -32.10, z-score = -2.60, R) >droEre2.scaffold_4690 17630083 106 - 18748788 -GGAAAUGGCUUUUUUGUUACAUUUUUUAUGAGCCCUCAAA--UUCCCUCGGAUUUAUUAAUGUAACAGAGAAGUGCUCAAUUAUGG-----CCGAGUCCGUGCAAACGGAAUU -((..(((((((((((((((((((....((((..((.....--.......))..)))).)))))))))))))))).......)))..-----))...(((((....)))))... ( -28.21, z-score = -2.10, R) >droAna3.scaffold_13334 424090 98 + 1562580 AAGCUUCAGUUUUUAUUAUAAAUAUUAUACAAGACCACCCC-CUGCCCCCUGCCCUGAAAAUACUUCAAAAUUAUGCUCAAUUAUG---------------UUCAAAUGGAAUU .(((....(((((...((((.....)))).)))))......-.............((((.....)))).......))).......(---------------(((.....)))). ( -3.30, z-score = 2.31, R) >consensus _GGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCAAA__UUCCCUCGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGA_____CCAAGUCCGUGCAAACGGAAUU ........(((((((((((((.((....((((((((..............)))))))).)).)))))))))))))......................(((((....)))))... (-17.10 = -18.56 + 1.45)
| Location | 21,099,866 – 21,099,956 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.34 |
| Shannon entropy | 0.13139 |
| G+C content | 0.32808 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.31 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21099866 90 + 22422827 CUUCUCUGUUACGUUAAUAAAUCCGAGGGAACUUUUGAGGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUGCUUAAUUUGCAAAUAGUU ......(((((((((...........(((..((....))..)))......)))))))))....(((.((((((.......)))))).))) ( -16.93, z-score = -1.51, R) >droSim1.chrX 16269767 88 + 17042790 CUUCUCUGUUACGUUAAUAAAUCCAAGGCAACUU--GAGGACCCAUCAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUU ......(((((((((........((((....)))--).(....)......)))))))))....(((.((((((.......)))))).))) ( -20.20, z-score = -3.33, R) >droSec1.super_8 3406528 88 + 3762037 CUUCUCUGUUACGUUAAUAAGUCCGAGGGAACUU--GAGGACCCAUCAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUU ......(((((((((.....(((((((....)))--..))))........)))))))))....(((.((((((.......)))))).))) ( -20.62, z-score = -2.23, R) >droYak2.chrX 20272070 88 + 21770863 CUUCUCUGUUACAUUAAUAAAUCCGAGGGGAUUU--GAAGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUCCCUAAUUUGGAAAUAGUU ......(((((((((...((((((....))))))--(......)......)))))))))....(((.((((((.......)))))).))) ( -17.20, z-score = -1.55, R) >droEre2.scaffold_4690 17630118 88 + 18748788 CUUCUCUGUUACAUUAAUAAAUCCGAGGGAAUUU--GAGGGCUCAUAAAAAAUGUAACAAAAAAGCCAUUUCCCUAAUUUGGAAAUAGUU ......(((((((((......(((...)))...(--((....))).....)))))))))....(((.((((((.......)))))).))) ( -15.40, z-score = -0.66, R) >consensus CUUCUCUGUUACGUUAAUAAAUCCGAGGGAACUU__GAGGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUU ......(((((((((...........(((..((....))..)))......)))))))))....(((.((((((.......)))))).))) (-15.23 = -14.31 + -0.92)
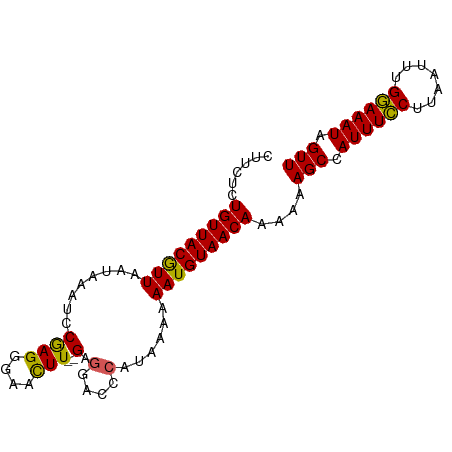
| Location | 21,099,866 – 21,099,956 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.34 |
| Shannon entropy | 0.13139 |
| G+C content | 0.32808 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.56 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
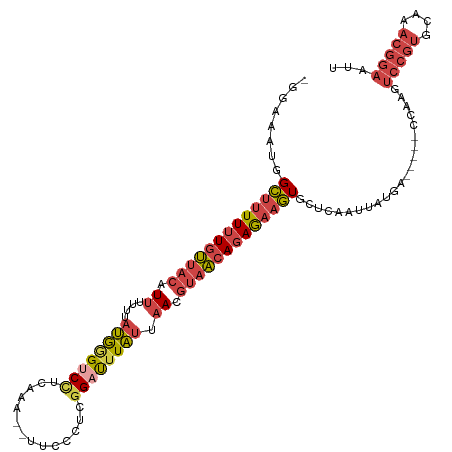
>dm3.chrX 21099866 90 - 22422827 AACUAUUUGCAAAUUAAGCAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCAAAAGUUCCCUCGGAUUUAUUAACGUAACAGAGAAG ..((((((((.......))))))))((((((((((((.((....((((((((..............)))))))).)).)))))))))))) ( -24.94, z-score = -4.13, R) >droSim1.chrX 16269767 88 - 17042790 AACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUC--AAGUUGCCUUGGAUUUAUUAACGUAACAGAGAAG ..((((((((.......))))))))((((((((((((....(((((((((((..--..........))))))))))).)))))))))))) ( -28.60, z-score = -4.79, R) >droSec1.super_8 3406528 88 - 3762037 AACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUC--AAGUUCCCUCGGACUUAUUAACGUAACAGAGAAG ..((((((((.......))))))))((((((((((((....(((((((((((..--..........))))))))))).)))))))))))) ( -30.40, z-score = -5.29, R) >droYak2.chrX 20272070 88 - 21770863 AACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCUUC--AAAUCCCCUCGGAUUUAUUAAUGUAACAGAGAAG ..((((((((.......))))))))(((((((((((((((....((((((((..--..........)))))))).))))))))))))))) ( -27.00, z-score = -4.26, R) >droEre2.scaffold_4690 17630118 88 - 18748788 AACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGAGCCCUC--AAAUUCCCUCGGAUUUAUUAAUGUAACAGAGAAG ..((((((((.......))))))))(((((((((((((((....((((..((..--..........))..)))).))))))))))))))) ( -25.60, z-score = -4.33, R) >consensus AACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUC__AAGUUCCCUCGGAUUUAUUAACGUAACAGAGAAG ..((((((((.......))))))))((((((((((((.((....((((((((..............)))))))).)).)))))))))))) (-24.90 = -24.30 + -0.60)

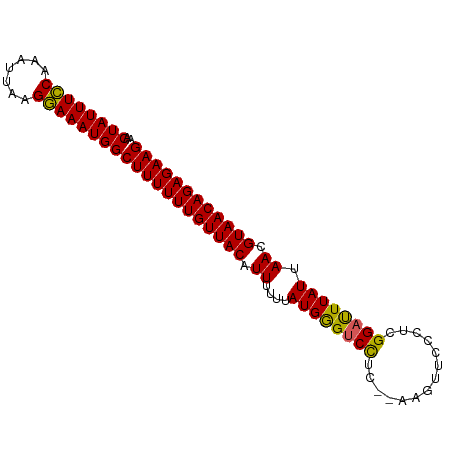
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:03 2011