
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,085,816 – 21,085,886 |
| Length | 70 |
| Max. P | 0.507531 |

| Location | 21,085,816 – 21,085,886 |
|---|---|
| Length | 70 |
| Sequences | 12 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Shannon entropy | 0.04710 |
| G+C content | 0.36100 |
| Mean single sequence MFE | -13.77 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.16 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507531 |
| Prediction | RNA |
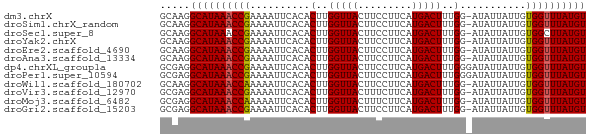
Download alignment: ClustalW | MAF
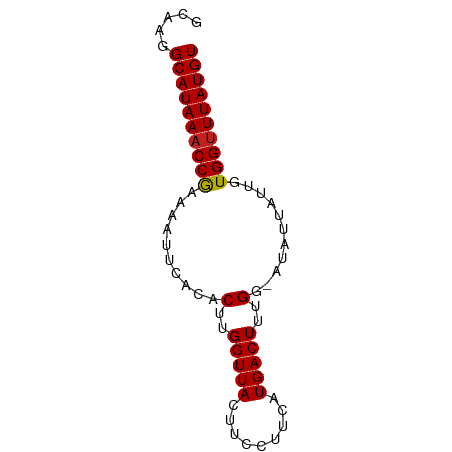
>dm3.chrX 21085816 70 + 22422827 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.12, R) >droSim1.chrX_random 5456546 70 + 5698898 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.12, R) >droSec1.super_8 3393701 70 + 3762037 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGCUUAUGU ..(((.((((((((((..........))))))...(((...........))-)......)))).))).... ( -11.00, z-score = -0.04, R) >droYak2.chrX 20257306 70 + 21770863 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.12, R) >droEre2.scaffold_4690 17616530 70 + 18748788 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.12, R) >droAna3.scaffold_13334 411990 70 - 1562580 GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.12, R) >dp4.chrXL_group1a 9129 71 - 9151740 GCGAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGGGAUAUUAUUGUGGUUUAUGU .....(((((((((((....))(((..((((....((((..........)))).)))).)))))))))))) ( -14.10, z-score = -0.73, R) >droPer1.super_10594 638 71 - 1032 GCGAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGGGAUAUUAUUGUGGUUUAUGU .....(((((((((((....))(((..((((....((((..........)))).)))).)))))))))))) ( -14.10, z-score = -0.73, R) >droWil1.scaffold_180702 72954 70 + 4511350 GCAAGGCAUAAACCAAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -14.55, z-score = -1.43, R) >droVir3.scaffold_12970 7823751 70 + 11907090 GCGAGGCAUAAACCGAAAAUUCACACUUGGUUACUUUCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -1.32, R) >droMoj3.scaffold_6482 1016570 70 - 2735782 GCGAGGCAUAAACCAAAAAUUCACACUUGGUUACUUUCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -14.55, z-score = -1.73, R) >droGri2.scaffold_15203 789617 70 - 11997470 GCGAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG-AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..).-.........)))))))))) ( -13.85, z-score = -0.96, R) >consensus GCAAGGCAUAAACCGAAAAUUCACACUUGGUUACUUCCUUCAUGACUUUGG_AUAUUAUUGUGGUUUAUGU .....((((((((((..........(..(((((.........)))))..)...........)))))))))) (-13.21 = -13.16 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:06:00 2011