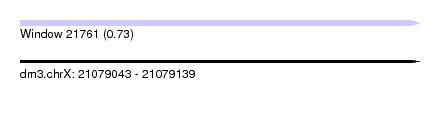
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,079,043 – 21,079,139 |
| Length | 96 |
| Max. P | 0.732877 |

| Location | 21,079,043 – 21,079,139 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.93 |
| Shannon entropy | 0.66551 |
| G+C content | 0.49650 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -7.45 |
| Energy contribution | -7.71 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
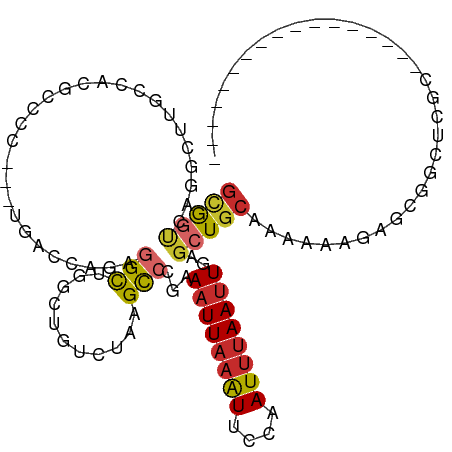
>dm3.chrX 21079043 96 + 22422827 GCGGUCAGGCUUGCCACGCCCCUC-UGACCAGAGGCUGUCUGUCUAAGCCUGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCGGCUCCC----------------- ....(((((((((.((.((.((((-(....)))))..)).))..)))))))))((((((((....))))))))(((((((........)))))))..----------------- ( -35.10, z-score = -3.64, R) >droSim1.chrX_random 5450719 97 + 5698898 GCGGUCAGGCUUGCCACGCCCCCCCUGACCAGAGGCUGGCUGGCUAAGCCUGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCGGCUCGC----------------- ((..((((((((((((.(((..((((....)).))..))))))).))))))))((((((((....))))))))(((((((........)))))))))----------------- ( -43.30, z-score = -4.39, R) >droSec1.super_8 3386900 97 + 3762037 GCGGUCAGGCUUGCCACGCCCCCCCUGACCAGAGGCUGGCUGGCUAAGCCUGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAUAAGAGCGGCUCGC----------------- ((..((((((((((((.(((..((((....)).))..))))))).))))))))((((((((....))))))))(((((((........)))))))))----------------- ( -43.30, z-score = -4.40, R) >droYak2.chrX 20250529 102 + 21770863 GUGGUCAGACUUGCCACGCCCCCUCUGACCAGAGGCUGGCUGUCUAAGCCCGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCGGUUCGGUUCCC------------ (((((.......)))))(((.(((((....)))))..)))......((((...((((((((....))))))))(((((((........)))))))))))...------------ ( -32.50, z-score = -2.40, R) >droEre2.scaffold_4690 17609751 97 + 18748788 GCGGUCAGACUUGCCACGCCCCCGCUGACCAGAGGCUGGCUGUCUAAGCCCGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCGGCUCCC----------------- .(((.(((((..((((.(((....((....)).))))))).))))..).))).((((((((....))))))))(((((((........)))))))..----------------- ( -31.90, z-score = -2.44, R) >droAna3.scaffold_13334 402113 83 - 1562580 GUGGUCAGACU--------------UGACCGGAGGCUUCCUGUCUAAGCCCGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCAACCAGC----------------- ..((((.....--------------.))))((..((((........(((....((((((((....))))))))..)))........))))..))...----------------- ( -19.49, z-score = -1.10, R) >droVir3.scaffold_12970 7817944 101 + 11907090 GCACCUAAGUCUGUGGC--------UG-UGGCACGCACUGGCUCUACGUCCCAAUUUAAGUUACAAUACAAUUGGCAUAAAAAAAACAGAGAGUA----GUGCAACGUGCCUUC (((((.........)).--------))-)(((((((((((.((((..((.((((((...((......))))))))..........))..))))))----))))...)))))... ( -24.00, z-score = -0.07, R) >droMoj3.scaffold_6482 1010534 106 - 2735782 GCAACUAAGUCUGCGGC--------UGAUGUCCGGUACUCGCUCUAAGCCUGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAACAUGGAGCAACGAGUGCAACGAGCCUUC (((........)))(((--------(..(((...(((((((((((((((....((((((((....))))))))..)))..........))))))...)))))).)))))))... ( -26.70, z-score = -1.09, R) >droGri2.scaffold_15203 782283 101 - 11997470 GUAGCAAGGAUUGCGGC--------UGUUGUC-UGCACUGGGGGCAAGCCCCAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAACAGAGAGAA----AUGCAACCGGCCUUC ...((..((.(((((.(--------((((...-((((((.((((....)))).((((((((....)))))))).)).))))...)))))......----.))))))).)).... ( -28.20, z-score = -0.59, R) >consensus GCGGUCAGGCUUGCCACGCCCC___UGACCAGAGGCUGGCUGUCUAAGCCCGAAAUUAAAUUCCAAUUUAAUUGAGCUGCAAAAAAGAGCGGCUCGC_________________ (((((............................(((...........)))...((((((((....))))))))..))))).................................. ( -7.45 = -7.71 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:57 2011