
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,257,641 – 12,257,733 |
| Length | 92 |
| Max. P | 0.876982 |

| Location | 12,257,641 – 12,257,733 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 68.05 |
| Shannon entropy | 0.48274 |
| G+C content | 0.47809 |
| Mean single sequence MFE | -13.51 |
| Consensus MFE | -8.19 |
| Energy contribution | -7.87 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12257641 92 + 23011544 UUUUGUUGUUC-GAGGGAAGCCCAUAAUUAUGAUUAUGAGUACUCCCUCAGAAUUUUCCUGCCGCCCCCCCCCCCCCUCCCCCUUCCCACCAU .......((((-((((((.((.((((((....)))))).))..)))))).))))....................................... ( -16.50, z-score = -2.69, R) >droSim1.chr2L 12059409 70 + 22036055 UUUUGUUGUUC-GAGGGAAGCCCAUAAUUAUGAUUAUGAGUACUCCCCCAGCAUUUUCCUGCCCCCCGCCC---------------------- ......((((.-(.((((.((.((((((....)))))).))..))))).))))..................---------------------- ( -12.80, z-score = -1.37, R) >droSec1.super_16 447813 70 + 1878335 UUUUGUUGUUC-GAGGGAAGCCCAUAAUUAUGAUUAUGAGUACUCCCCCAGCAUUUUCCUGCCCCCCGCCC---------------------- ......((((.-(.((((.((.((((((....)))))).))..))))).))))..................---------------------- ( -12.80, z-score = -1.37, R) >dp4.chr4_group3 5815173 79 - 11692001 UUUUGCUGUGAAGAAGGAAGCACAUAAUUAUGAUUAUG-GUGCCUCUAUAUCUCUCUCUUUCUGCCCAUCAUCAUUGUUG------------- ...((((...........))))..((((.(((((.(((-(.((....................)))))).))))).))))------------- ( -11.95, z-score = 0.55, R) >consensus UUUUGUUGUUC_GAGGGAAGCCCAUAAUUAUGAUUAUGAGUACUCCCCCAGCAUUUUCCUGCCCCCCGCCC______________________ ..............((((.((.((((((....)))))).))..)))).............................................. ( -8.19 = -7.87 + -0.31)


| Location | 12,257,641 – 12,257,733 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 68.05 |
| Shannon entropy | 0.48274 |
| G+C content | 0.47809 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
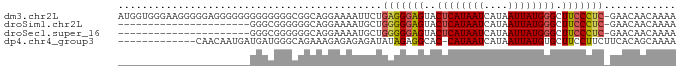
>dm3.chr2L 12257641 92 - 23011544 AUGGUGGGAAGGGGGAGGGGGGGGGGGGGCGGCAGGAAAAUUCUGAGGGAGUACUCAUAAUCAUAAUUAUGGGCUUCCCUC-GAACAACAAAA .((.((....((((((((...........(..(((((...)))))..).....((((((((....))))))))))))))))-...)).))... ( -23.50, z-score = -1.09, R) >droSim1.chr2L 12059409 70 - 22036055 ----------------------GGGCGGGGGGCAGGAAAAUGCUGGGGGAGUACUCAUAAUCAUAAUUAUGGGCUUCCCUC-GAACAACAAAA ----------------------(..((...((((......))))(((((((..((((((((....))))))))))))))))-)..)....... ( -22.00, z-score = -2.65, R) >droSec1.super_16 447813 70 - 1878335 ----------------------GGGCGGGGGGCAGGAAAAUGCUGGGGGAGUACUCAUAAUCAUAAUUAUGGGCUUCCCUC-GAACAACAAAA ----------------------(..((...((((......))))(((((((..((((((((....))))))))))))))))-)..)....... ( -22.00, z-score = -2.65, R) >dp4.chr4_group3 5815173 79 + 11692001 -------------CAACAAUGAUGAUGGGCAGAAAGAGAGAGAUAUAGAGGCAC-CAUAAUCAUAAUUAUGUGCUUCCUUCUUCACAGCAAAA -------------......................(((..((.....(((((((-.(((((....))))))))))))))..)))......... ( -13.30, z-score = -0.03, R) >consensus ______________________GGGCGGGCGGCAGGAAAAUGCUGGGGGAGUACUCAUAAUCAUAAUUAUGGGCUUCCCUC_GAACAACAAAA ............................................((((((((...((((((....)))))).))))))))............. (-11.35 = -11.35 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:40 2011