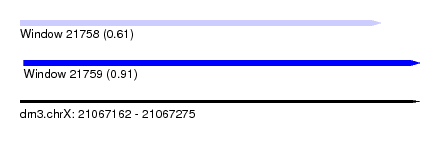
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,067,162 – 21,067,275 |
| Length | 113 |
| Max. P | 0.906439 |

| Location | 21,067,162 – 21,067,264 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Shannon entropy | 0.34967 |
| G+C content | 0.48322 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
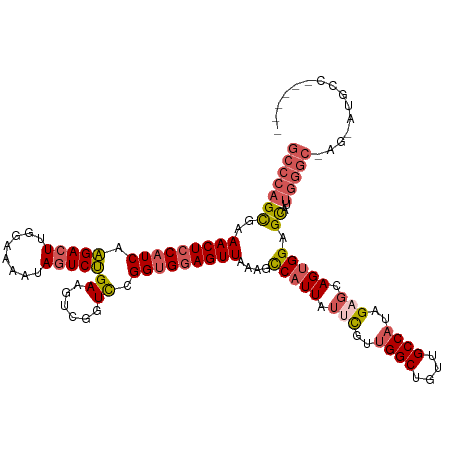
>dm3.chrX 21067162 102 + 22422827 ------------------AGCCCAGCAAAACUCCAUCAAGACUUGGGAAAUAGUCUGAAGUCGGUCCGGUGGAGUUAAAGCCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUU ------------------......((..(((((((((..(((((.(((.....))).))))).....)))))))))....(((((.(((..((((....))))..))).))))).))... ( -37.00, z-score = -3.30, R) >droEre2.scaffold_4690 17598114 103 + 18748788 ----------------CUGGCCCAGCAAAACUGCAUCAGGACUUGGCAGGUAGUCCGAAGUCGGUCCGGUGGAGUUAAAGCCCUUCUUCGU-GGCUGUUGCCAUAGUGCAGUGGAGCAUU ----------------...((((((((.((((.((((.(((((.(((.((....))...)))))))))))).)))).............((-(((....)))))..)))..))).))... ( -34.70, z-score = -0.39, R) >droYak2.chrX 20238131 98 + 21770863 ----------------------CUGUGAAACUCCAUCAAGACUCGGGAAAUAUUCUGAAUUGGGUUCGGAGGAGUUAAAGCCAUUAUUUGCUGGCUGUUGCCAUAGUGCAGUGGAGUAGU ----------------------(((((.((((((.((..(((((((.............)))))))..))))))))..(((((........))))).....)))))(((......))).. ( -24.82, z-score = -0.12, R) >droSec1.super_8 3375309 120 + 3762037 CUAUGAAAUUGCGCCUCCAGCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUUGGUCCGGUGGAGUUAAAGUCAUUAUUCGUUGGCUGUUGCCAUAGACCAGUGGAGCAUU .........(((.....(((((.((((((((((((((.(((((........)))))((......)).))))))))...........)))))))))))...((((......)))).))).. ( -34.20, z-score = -1.26, R) >droSim1.chrX 16247940 120 + 17042790 CUAUGAAAUUGCGCCUCCAGCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUUGGUCCGGUGGAGUUAAAGUCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUU .........(((.((..(((((.((((((((((((((.(((((........)))))((......)).))))))))...........)))))))))))((((......)))).)).))).. ( -35.70, z-score = -1.51, R) >consensus ________________C_AGCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUCGGUCCGGUGGAGUUAAAGCCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUU ........................((..(((((((((.(((((........)))))((......)).)))))))))....(((((.(((..((((....))))..))).))))).))... (-23.86 = -24.26 + 0.40)

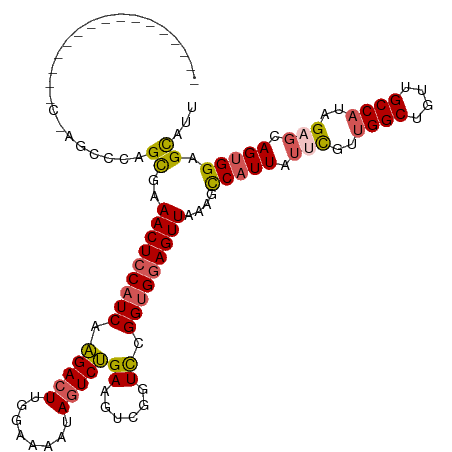
| Location | 21,067,163 – 21,067,275 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Shannon entropy | 0.30152 |
| G+C content | 0.50594 |
| Mean single sequence MFE | -39.46 |
| Consensus MFE | -27.60 |
| Energy contribution | -29.00 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21067163 112 + 22422827 GCCCAGCAAAACUCCAUCAAGACUUGGGAAAUAGUCUGAAGUCGGUCCGGUGGAGUUAAAGCCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUUGGGC-AG-AUGCC------ (((((((..(((((((((..(((((.(((.....))).))))).....)))))))))....(((((.(((..((((....))))..))).))))).))..)))))-..-.....------ ( -45.60, z-score = -3.96, R) >droEre2.scaffold_4690 17598117 116 + 18748788 GCCCAGCAAAACUGCAUCAGGACUUGGCAGGUAGUCCGAAGUCGGUCCGGUGGAGUUAAAGCCCUUCUUCGU-GGCUGUUGCCAUAGUGCAGUGGAGCAUUGGGCCAG-GAGAAGAUG-- .....((..((((.((((.(((((.(((.((....))...)))))))))))).))))...)).(((((((((-(((....))))).((.(((((...))))).))..)-))))))...-- ( -41.50, z-score = -0.93, R) >droYak2.chrX 20238133 115 + 21770863 -----GUGAAACUCCAUCAAGACUCGGGAAAUAUUCUGAAUUGGGUUCGGAGGAGUUAAAGCCAUUAUUUGCUGGCUGUUGCCAUAGUGCAGUGGAGUAGUGGAGUAGUGGGCAGAUGCC -----....((((((.((..(((((((.............)))))))..))))))))....((((((((..(((.(..((((......))))..)..)))..)).))))))((....)). ( -33.82, z-score = -1.01, R) >droSec1.super_8 3375328 112 + 3762037 GCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUUGGUCCGGUGGAGUUAAAGUCAUUAUUCGUUGGCUGUUGCCAUAGACCAGUGGAGCAUUGGGC-AG-AUGCC------ ((((((.(...((((((((.((((........)))))))..((((((..((((......(((((........)))))....)))).)))))))))))).))))))-..-.....------ ( -38.00, z-score = -2.27, R) >droSim1.chrX 16247959 112 + 17042790 GCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUUGGUCCGGUGGAGUUAAAGUCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUUGGGC-AG-AUGCC------ (((((((..(((((((((.(((((........)))))((......)).)))))))))....(((((.(((..((((....))))..))).))))).))..)))))-..-.....------ ( -38.40, z-score = -2.36, R) >consensus GCCCAGCGAAACUCCAUCAAGACUUGGAAAAUAGUCUGAAGUCGGUCCGGUGGAGUUAAAGCCAUUAUUCGUUGGCUGUUGCCAUAGAGCAGUGGAGCAUUGGGC_AG_AUGCC______ (((((((..(((((((((.(((((........)))))((......)).)))))))))....(((((.(((..((((....))))..))).))))).))..)))))............... (-27.60 = -29.00 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:56 2011