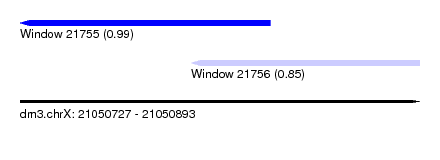
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,050,727 – 21,050,893 |
| Length | 166 |
| Max. P | 0.988004 |

| Location | 21,050,727 – 21,050,831 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Shannon entropy | 0.31429 |
| G+C content | 0.45759 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.988004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
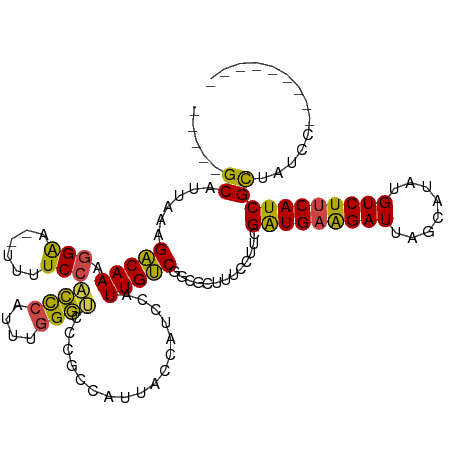
>dm3.chrX 21050727 104 - 22422827 -----GCAUUAAAGACAAAGGAA--UUUUCCACCCAUUUGGGUUCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC--------- -----((..........((((((--......((((....))))..((((................))))..)))))).(((((((((........))))))))))).....--------- ( -28.39, z-score = -3.29, R) >droAna3.scaffold_13334 1200486 120 - 1562580 AAUUGGCAUUAAAGUCAAAAGGAGCUACUCGUUUCCUCCUGAACUGAUGGCUGCCAUCCAUUGACGAUGGUUACCUUCGGUGUGGAUUAGCAUAUGUCUUCAUCGUCCUGGUUGCACUCC ..(((((......)))))..((((......(((...(((...(((((.((..((((((.......))))))..)).)))))..)))..)))...(((..(((......)))..))))))) ( -32.30, z-score = -0.88, R) >droEre2.scaffold_4690 17583216 104 - 18748788 -----GCAUUAAAGACAAAGGAA--UUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC--------- -----((......(((((.(((.--......((((....))))...((....))..))).))))).............(((((((((........))))))))))).....--------- ( -28.40, z-score = -2.54, R) >droYak2.chrX 20219734 104 - 21770863 -----GCAUUAAAGACAAAGGAA--UUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCAUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC--------- -----((......(((((.(((.--......((((....))))...((....))..))).))))).............(((((((((........))))))))))).....--------- ( -28.40, z-score = -2.47, R) >droSec1.super_8 3358640 104 - 3762037 -----GCAUUAAAGACAAAGGAA--UUUUCCUCCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGGUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC--------- -----((..........(((((.--.......(((....)))...((((................))))...))))).(((((((((........))))))))))).....--------- ( -25.59, z-score = -1.82, R) >droSim1.chrX 16226555 104 - 17042790 -----GCAUUAAAGACAAAGGAA--UUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC--------- -----((..........(((((.--......((((....))))..((((................))))...))))).(((((((((........))))))))))).....--------- ( -27.59, z-score = -2.93, R) >consensus _____GCAUUAAAGACAAAGGAA__UUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCC_________ .....((......(((((.((((....))))((((....)))).................))))).............(((((((((........))))))))))).............. (-23.80 = -23.08 + -0.72)

| Location | 21,050,798 – 21,050,893 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Shannon entropy | 0.14491 |
| G+C content | 0.35439 |
| Mean single sequence MFE | -17.20 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21050798 95 - 22422827 ACUAAGCUUUUUUUUUUUAAAUGUCACCCCUCAAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUG .....................((((..((.((((((......)))))).))..(((((....)))))...))))..(((....)))......... ( -16.90, z-score = -1.76, R) >droEre2.scaffold_4690 17583287 92 - 18748788 ACUAAGC---AUUUUUUAAAAUGUCACCCUUCCUUCCACAUUAGUUGAUGGAAAACGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUG .......---...........((((....((((.((.((....)).)).))))...((....))......))))..(((....)))......... ( -14.00, z-score = -1.11, R) >droYak2.chrX 20219805 92 - 21770863 ACUAAGC---AUAUUUUAAAAUGUCAGUCUUCCUUCCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUG .......---........(((((...(((((..(((((((.....)).)))))(((((....))))).)))))...(((....)))...))))). ( -18.40, z-score = -2.07, R) >droSec1.super_8 3358711 92 - 3762037 ACUAAGC---UUUUUUCAAGAUGUCGCCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCUCCCAUUUG .......---...........((((.....((((((.((....)).)))))).(((((....)))))...)))).((((....))))........ ( -18.40, z-score = -1.72, R) >droSim1.chrX 16226626 92 - 17042790 ACUAAGC---UUUUUUCAAGAUGUCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUG .......---...........((((.....((((((.((....)).)))))).(((((....)))))...))))..(((....)))......... ( -18.30, z-score = -2.00, R) >consensus ACUAAGC___UUUUUUUAAAAUGUCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUG .....................((((.....((((((.((....)).)))))).(((((....)))))...))))..(((....)))......... (-15.36 = -15.92 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:53 2011