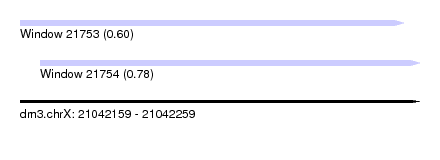
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,042,159 – 21,042,259 |
| Length | 100 |
| Max. P | 0.780029 |

| Location | 21,042,159 – 21,042,255 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.42776 |
| G+C content | 0.39603 |
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -8.56 |
| Energy contribution | -9.80 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
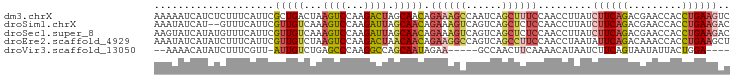
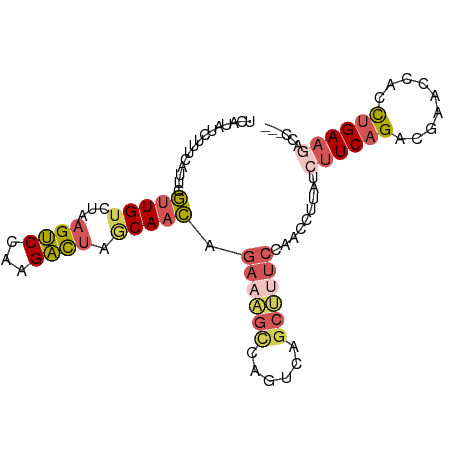
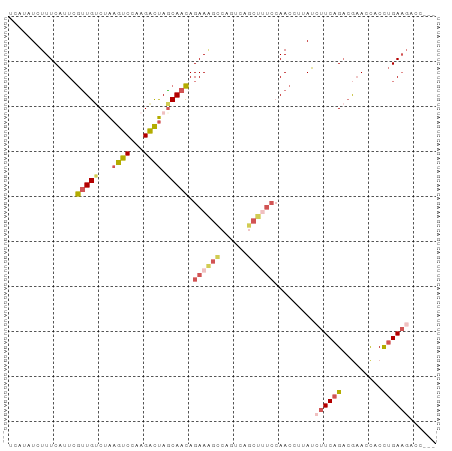
>dm3.chrX 21042159 96 + 22422827 AAAAAUCAUCUCUUUCAUUCGCUGACUAAGUCCAAGACUAGCAACAGAAAGCCAAUCAGCUUUCCAACCUUAUCUUCAGACGAACCACCUGAAGUC ............(((((((((((((...((((...)))).......((((((......))))))...........)))).)))).....))))).. ( -16.50, z-score = -2.00, R) >droSim1.chrX 16165119 94 + 17042790 AAAUAUCAU--GUUUCAUUCGUUGUCAAAGUCCAAGAUUAGCAACAGAAAGUCAGUCAGCUCUCCAACCUUAUCUUCAGACGAACCACCUGAAGAC .........--.........(((((...((((...)))).))))).((.(((......))).))........(((((((.........))))))). ( -12.80, z-score = -0.00, R) >droSec1.super_8 412837 96 - 3762037 AAGUAUCAUAUGUUUCAUUCGUUGUCAAAGUCCAAGAUUAGCAACAGAAAGUCAGUCAGCUCUCCAACCUUAUCUUCAGACGAACCACCUGAAGAC ....................(((((...((((...)))).))))).((.(((......))).))........(((((((.........))))))). ( -12.80, z-score = 0.26, R) >droEre2.scaffold_4929 751497 96 - 26641161 AAAUAUCAUAUCUUUCAUUCGUUGUCUAAGUCCAAGACUAACAACAGAAGGCCAGUCAGCCUUCCAACCUAAUAUUCAGACAAACCACCUGAAGCU ....................(((((...((((...)))).))))).((((((......))))))..........(((((.........)))))... ( -18.20, z-score = -3.18, R) >droVir3.scaffold_13050 2046783 84 + 3426691 --AAAACAUAUCUUUCGUU-AUUGUCUGAGCCCAAGGCCAGCAAUAGAA-----GCCAACUUCAAAACAUAAUCUUCAGUAAUAUUACUGGA---- --................(-((((.(((.(((...)))))))))))(((-----(....))))...........(((((((....)))))))---- ( -17.80, z-score = -2.72, R) >consensus AAAUAUCAUAUCUUUCAUUCGUUGUCUAAGUCCAAGACUAGCAACAGAAAGCCAGUCAGCUUUCCAACCUUAUCUUCAGACGAACCACCUGAAGAC ....................(((((...((((...)))).))))).((((((......)))))).........((((((.........)))))).. ( -8.56 = -9.80 + 1.24)
| Location | 21,042,164 – 21,042,259 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 74.41 |
| Shannon entropy | 0.45850 |
| G+C content | 0.42116 |
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -8.56 |
| Energy contribution | -9.80 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
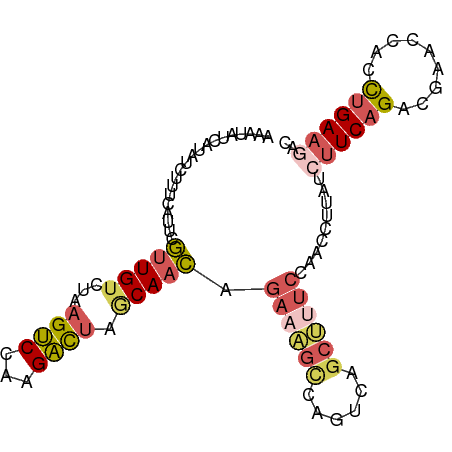
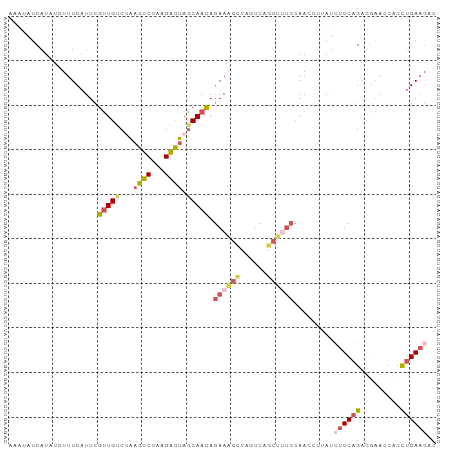
>dm3.chrX 21042164 95 + 22422827 UCAUCUCUUUCAUUCGCUGACUAAGUCCAAGACUAGCAACAGAAAGCCAAUCAGCUUUCCAACCUUAUCUUCAGACGAACCACCUGAAGUCCCAC .......(((((((((((((...((((...)))).......((((((......))))))...........)))).)))).....)))))...... ( -16.50, z-score = -2.17, R) >droSim1.chrX 16165124 90 + 17042790 --UCAUGUUUCAUUCGUUGUCAAAGUCCAAGAUUAGCAACAGAAAGUCAGUCAGCUCUCCAACCUUAUCUUCAGACGAACCACCUGAAGACC--- --.............(((((...((((...)))).))))).((.(((......))).))........(((((((.........)))))))..--- ( -12.80, z-score = 0.01, R) >droSec1.super_8 412842 92 - 3762037 UCAUAUGUUUCAUUCGUUGUCAAAGUCCAAGAUUAGCAACAGAAAGUCAGUCAGCUCUCCAACCUUAUCUUCAGACGAACCACCUGAAGACC--- ...............(((((...((((...)))).))))).((.(((......))).))........(((((((.........)))))))..--- ( -12.80, z-score = -0.07, R) >droEre2.scaffold_4929 751502 94 - 26641161 UCAUAUCUUUCAUUCGUUGUCUAAGUCCAAGACUAACAACAGAAGGCCAGUCAGCCUUCCAACCUAAUAUUCAGACAAACCACCUGAAGCUCAC- ...............(((((...((((...)))).))))).((((((......))))))..........(((((.........)))))......- ( -18.20, z-score = -3.04, R) >droVir3.scaffold_13050 2046786 81 + 3426691 ACAUAUCUUUCGUU-AUUGUCUGAGCCCAAGGCCAGCAAUAGAA-----GCCAACUUCAAAACAUAAUCUUCAGUAAUAUUACUGGA-------- .............(-((((.(((.(((...)))))))))))(((-----(....))))...........(((((((....)))))))-------- ( -17.80, z-score = -2.69, R) >consensus UCAUAUCUUUCAUUCGUUGUCUAAGUCCAAGACUAGCAACAGAAAGCCAGUCAGCUUUCCAACCUUAUCUUCAGACGAACCACCUGAAGACC___ ...............(((((...((((...)))).))))).((((((......)))))).........((((((.........))))))...... ( -8.56 = -9.80 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:52 2011